1C9A
 
 | | SOLUTION STRUCTURE OF NEUROMEDIN B | | Descriptor: | NEUROMEDIN B | | Authors: | Lee, S, Kim, Y. | | Deposit date: | 1999-08-01 | | Release date: | 1999-11-11 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of neuromedin B by (1)H nuclear magnetic resonance spectroscopy.
FEBS Lett., 460, 1999
|
|
3R4Z
 
 | | Crystal structure of alpha-neoagarobiose hydrolase (ALPHA-NABH) in complex with alpha-d-galactopyranose from Saccharophagus degradans 2-40 | | Descriptor: | Glycosyl hydrolase family 32, N terminal, alpha-D-galactopyranose | | Authors: | Lee, S, Lee, J.Y, Ha, S.C, Shin, D.H, Kim, K.H, Bang, W.G, Kim, S.H, Choi, I.G. | | Deposit date: | 2011-03-18 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of a key enzyme in the agarolytic pathway, alpha-neoagarobiose hydrolase from Saccharophagus degradans 2-40
Biochem.Biophys.Res.Commun., 412, 2011
|
|
1C98
 
 | | SOLUTION STRUCTURE OF NEUROMEDIN B | | Descriptor: | NEUROMEDIN B | | Authors: | Lee, S, Kim, Y. | | Deposit date: | 1999-08-01 | | Release date: | 1999-08-11 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of neuromedin B by (1)H nuclear magnetic resonance spectroscopy.
FEBS Lett., 460, 1999
|
|
2R0O
 
 | |
1F4V
 
 | | CRYSTAL STRUCTURE OF ACTIVATED CHEY BOUND TO THE N-TERMINUS OF FLIM | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CHEMOTAXIS CHEY PROTEIN, FLAGELLAR MOTOR SWITCH PROTEIN, ... | | Authors: | Lee, S.Y, Cho, H.S, Pelton, J.G, Yan, D, Henderson, R.K, King, D, Huang, L.S, Kustu, S, Berry, E.A, Wemmer, D.E. | | Deposit date: | 2000-06-10 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of an activated response regulator bound to its target.
Nat.Struct.Biol., 8, 2001
|
|
4DWZ
 
 | | Crystal Structure of Ton_0340 | | Descriptor: | Hypothetical protein TON_0340, ZINC ION | | Authors: | Lee, S.G, Lee, K.H, Cha, S.S, Oh, B.H. | | Deposit date: | 2012-02-27 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Experimental phasing using zinc anomalous scattering
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4FC5
 
 | | Crystal Structure of Ton_0340 | | Descriptor: | Putative uncharacterized protein, ZINC ION | | Authors: | Lee, S.G, Lee, K.H, An, Y.J, Cha, S.S, Oh, B.H. | | Deposit date: | 2012-05-24 | | Release date: | 2012-09-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Experimental phasing using zinc anomalous scattering
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6OQA
 
 | | Crystal structure of CEP250 bound to FKBP12 in the presence of FK506-like novel natural product | | Descriptor: | (3R,4E,7E,10R,11S,12R,13S,16R,17R,24aS)-11,17-dihydroxy-10,12,16-trimethyl-3-[(2R)-1-phenylbutan-2-yl]-6,9,10,11,12,13,14,15,16,17,22,23,24,24a-tetradecahydro-3H-13,17-epoxypyrido[2,1-c][1,4]oxazacyclohenicosine-1,18,19(21H)-trione, 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ... | | Authors: | Lee, S.-J, Shigdel, U.K, Townson, S.A, Verdine, G.L. | | Deposit date: | 2019-04-26 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Genomic discovery of an evolutionarily programmed modality for small-molecule targeting of an intractable protein surface.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6O87
 
 | |
6WLF
 
 | |
3DAK
 
 | |
4YJ6
 
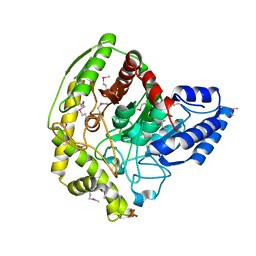 | | The Crystal Structure of a Bacterial Aryl Acylamidase Belonging to the Amidase signature (AS) enzymes family | | Descriptor: | Aryl acylamidase, PHOSPHATE ION | | Authors: | Lee, S, Park, E.-H, Ko, H.-J, Bang, W.-G, Choi, I.-G. | | Deposit date: | 2015-03-03 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure analysis of a bacterial aryl acylamidase belonging to the amidase signature enzyme family
Biochem.Biophys.Res.Commun., 467, 2015
|
|
5YI5
 
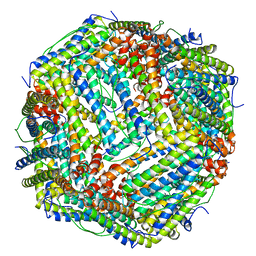 | | human ferritin mutant - E-helix deletion | | Descriptor: | Ferritin heavy chain | | Authors: | Lee, S.G, Yoon, H.R, Ahn, B.J, Jeong, H, Hyun, J, Jung, Y, Kim, H. | | Deposit date: | 2017-10-02 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Four-fold Channel-Nicked Human Ferritin Nanocages for Active Drug Loading and pH-Responsive Drug Release
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
7V6E
 
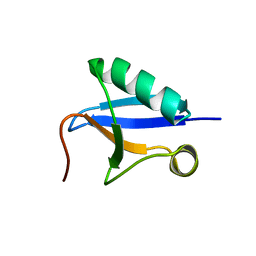 | | DREP3 | | Descriptor: | DNAation factor-related protein 3, isoform A | | Authors: | Lee, S.Y, Park, H.H. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Helical filament structure of the DREP3 CIDE domain reveals a unified mechanism of CIDE-domain assembly.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
1UKL
 
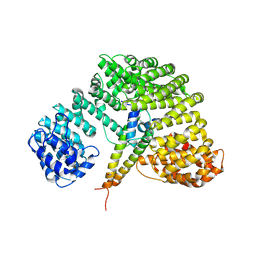 | | Crystal structure of Importin-beta and SREBP-2 complex | | Descriptor: | Importin beta-1 subunit, Sterol regulatory element binding protein-2 | | Authors: | Lee, S.J, Sekimoto, T, Yamashita, E, Nagoshi, E, Nakagawa, A, Imamoto, N, Yoshimura, M, Sakai, H, Tsukihara, T, Yoneda, Y. | | Deposit date: | 2003-08-26 | | Release date: | 2003-12-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of Importin-beta Bound to SREBP-2: Nuclear Import of a Transcription Factor
Science, 302, 2003
|
|
4HWL
 
 | | Crystal Structure Analysis of the Bacteriorhodopsin in Facial Amphiphile-7 DMPC Bicelle | | Descriptor: | Bacteriorhodopsin, GLYCEROL, HEPTANE, ... | | Authors: | Lee, S, Stout, C.D, Zhang, Q. | | Deposit date: | 2012-11-08 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Steroid-based facial amphiphiles for stabilization and crystallization of membrane proteins.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6MEV
 
 | | Structure of JMJD6 bound to Mono-Methyl Arginine. | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, 2-OXOGLUTARIC ACID, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, ... | | Authors: | Lee, S, Zhang, G. | | Deposit date: | 2018-09-07 | | Release date: | 2019-09-18 | | Last modified: | 2020-04-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | JMJD6 cleaves MePCE to release positive transcription elongation factor b (P-TEFb) in higher eukaryotes.
Elife, 9, 2020
|
|
5XB1
 
 | | human ferritin mutant - E-helix deletion | | Descriptor: | Ferritin heavy chain | | Authors: | Lee, S.G, Ahn, B.J, Jeong, H, Kim, H, Hyun, J, Jung, Y. | | Deposit date: | 2017-03-15 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Four-fold Channel-Nicked Human Ferritin Nanocages for Active Drug Loading and pH-Responsive Drug Release
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
4OMY
 
 | |
4YN4
 
 | |
3HEQ
 
 | | Human prion protein variant D178N with M129 | | Descriptor: | CADMIUM ION, Major prion protein | | Authors: | Lee, S, Antony, L, Hartmann, R, Knaus, K.J, Surewicz, K, Surewicz, W.K, Yee, V.C. | | Deposit date: | 2009-05-10 | | Release date: | 2010-01-12 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational diversity in prion protein variants influences intermolecular beta-sheet formation.
Embo J., 29, 2010
|
|
3HES
 
 | | Human prion protein variant F198S with M129 | | Descriptor: | CADMIUM ION, Major prion protein | | Authors: | Lee, S, Antony, L, Hartmann, R, Knaus, K.J, Surewicz, K, Surewicz, W.K, Yee, V.C. | | Deposit date: | 2009-05-10 | | Release date: | 2010-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational diversity in prion protein variants influences intermolecular beta-sheet formation.
Embo J., 29, 2010
|
|
4YMN
 
 | |
1FQW
 
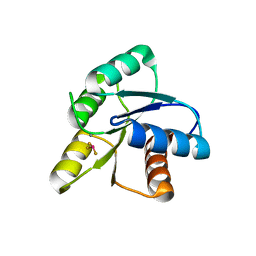 | | CRYSTAL STRUCTURE OF ACTIVATED CHEY | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CHEMOTAXIS CHEY PROTEIN, MANGANESE (II) ION | | Authors: | Lee, S.Y, Cho, H.S, Pelton, J.G, Yan, D, Berry, E.A, Wemmer, D.E. | | Deposit date: | 2000-09-07 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of activated CheY. Comparison with other activated receiver domains.
J.Biol.Chem., 276, 2001
|
|
4YMM
 
 | |
