4PKE
 
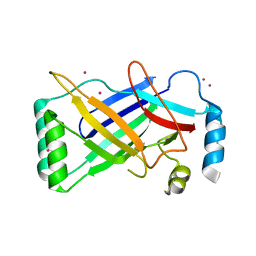 | | The structure of a conserved Piezo channel domain reveals a novel beta sandwich fold | | 分子名称: | PLATINUM (II) ION, Protein C10C5.1, isoform i | | 著者 | Kamajaya, A, Kaiser, J, Lee, J, Reid, M, Rees, D.C. | | 登録日 | 2014-05-14 | | 公開日 | 2014-10-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The Structure of a Conserved Piezo Channel Domain Reveals a Topologically Distinct beta Sandwich Fold.
Structure, 22, 2014
|
|
2GEF
 
 | |
2RVC
 
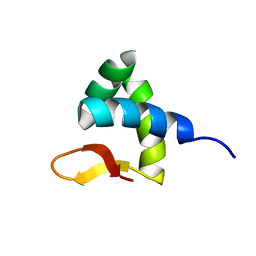 | | Solution structure of Zalpha domain of goldfish ZBP-containing protein kinase | | 分子名称: | Interferon-inducible and double-stranded-dependent eIF-2kinase | | 著者 | Lee, A, Park, C, Park, J, Kwon, M, Choi, Y, Kim, K, Choi, B, Lee, J. | | 登録日 | 2015-07-08 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the Z-DNA binding domain of PKR-like protein kinase from Carassius auratus and quantitative analyses of the intermediate complex during B-Z transition.
Nucleic Acids Res., 44, 2016
|
|
2VRC
 
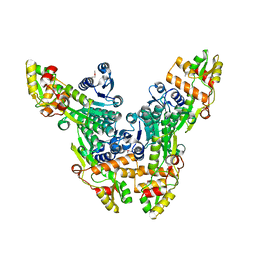 | | Crystal structure of the Citrobacter sp. triphenylmethane reductase complexed with NADP(H) | | 分子名称: | TRIPHENYLMETHANE REDUCTASE | | 著者 | Kim, Y, Park, H.J, Kwak, S.N, Lee, J.S, Oh, T.K, Kim, M.H. | | 登録日 | 2008-03-31 | | 公開日 | 2008-09-23 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Insight Into Bioremediation of Triphenylmethane Dyes by Citrobacter Sp. Triphenylmethane Reductase.
J.Biol.Chem., 283, 2008
|
|
2VRB
 
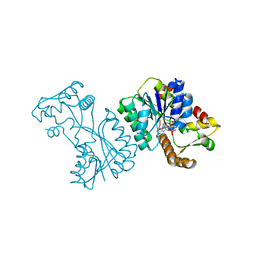 | | Crystal structure of the Citrobacter sp. triphenylmethane reductase complexed with NADP(H) | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRIPHENYLMETHANE REDUCTASE | | 著者 | Kim, Y, Park, H.J, Kwak, S.N, Lee, J.S, Oh, T.K, Kim, M.H. | | 登録日 | 2008-03-31 | | 公開日 | 2008-09-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Insight Into Bioremediation of Triphenylmethane Dyes by Citrobacter Sp. Triphenylmethane Reductase.
J.Biol.Chem., 283, 2008
|
|
6XPR
 
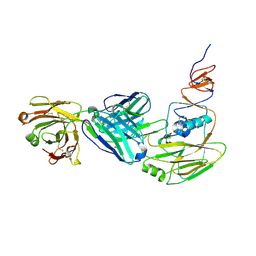 | | Human antibody D2 H1-1/H3-1 H3 in complex with the influenza hemagglutinin head domain of A/Texas/50/2012(H3N2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | 著者 | McCarthy, K.R, Harrison, S.C, Lee, J. | | 登録日 | 2020-07-08 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (4.092 Å) | | 主引用文献 | A Prevalent Focused Human Antibody Response to the Influenza Virus Hemagglutinin Head Interface.
Mbio, 12, 2021
|
|
6ZIF
 
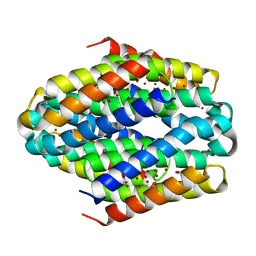 | |
8HU2
 
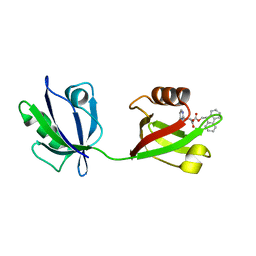 | |
6V05
 
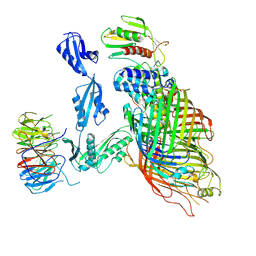 | | Cryo-EM structure of a substrate-engaged Bam complex | | 分子名称: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | 著者 | Tomasek, D, Rawson, S, Lee, J, Wzorek, J.S, Harrison, S.C, Li, Z, Kahne, D. | | 登録日 | 2019-11-18 | | 公開日 | 2020-06-10 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structure of a nascent membrane protein as it folds on the BAM complex.
Nature, 583, 2020
|
|
2BTN
 
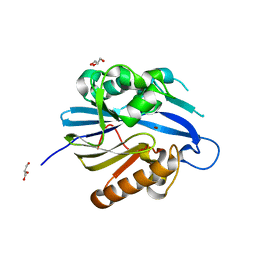 | | Crystal Structure and Catalytic Mechanism of the Quorum-Quenching N- Acyl Homoserine Lactone Hydrolase | | 分子名称: | AIIA-LIKE PROTEIN, GLYCEROL, ZINC ION | | 著者 | Kim, M.H, Choi, W.C, Kang, H.O, Lee, J.S, Kang, B.S, Kim, K.J, Derewenda, Z.S, Oh, T.K, Lee, C.H, Lee, J.K. | | 登録日 | 2005-06-03 | | 公開日 | 2005-12-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The Molecular Structure and Catalytic Mechanism of a Quorum-Quenching N-Acyl-L-Homoserine Lactone Hydrolase.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
8H26
 
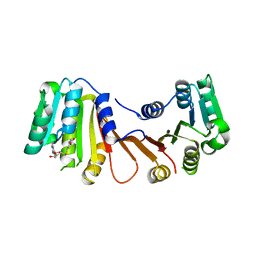 | |
8IS3
 
 | | Structural model for the micelle-bound indolicidin-like peptide in solution | | 分子名称: | Indolicidin-like antimicrobial peptide | | 著者 | Kim, B, Ko, Y.H, Kim, J, Lee, J, Nam, C.H, Kim, J.H. | | 登録日 | 2023-03-20 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural model for the micelle-bound indolicidin-like peptide in solution
To Be Published
|
|
8VBW
 
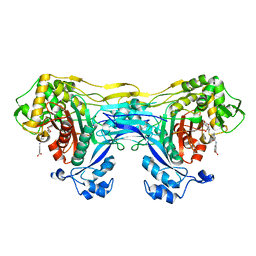 | | Structure of the monofunctional Staphylococcus aureus PBP1 in its beta-lactam (Ertapenem) inhibited form | | 分子名称: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein 1 | | 著者 | Bon, C.G, Lee, J, Caveney, N.A, Strynadka, N.C.J. | | 登録日 | 2023-12-12 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and kinetic analysis of the monofunctional Staphylococcus aureus PBP1.
J.Struct.Biol., 216, 2024
|
|
8VBV
 
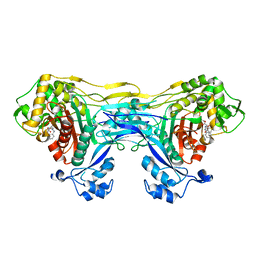 | | Structure of the monofunctional Staphylococcus aureus PBP1 in its beta-lactam (Cephalexin) inhibited form | | 分子名称: | (2S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5-methyl-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Penicillin-binding protein 1 | | 著者 | Bon, C.G, Lee, J, Caveney, N.A, Strynadka, N.C.J. | | 登録日 | 2023-12-12 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural and kinetic analysis of the monofunctional Staphylococcus aureus PBP1.
J.Struct.Biol., 216, 2024
|
|
8VBT
 
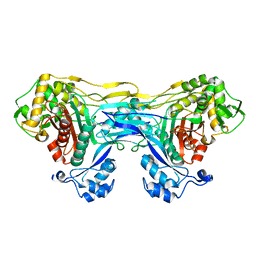 | |
8VBU
 
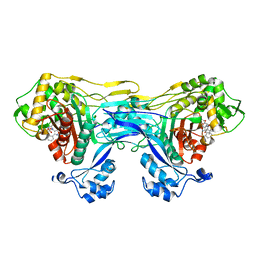 | | Structure of the monofunctional Staphylococcus aureus PBP1 in its beta-lactam (Oxacillin) inhibited form | | 分子名称: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, Penicillin-binding protein 1 | | 著者 | Bon, C.G, Lee, J, Caveney, N.A, Strynadka, N.C.J. | | 登録日 | 2023-12-12 | | 公開日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and kinetic analysis of the monofunctional Staphylococcus aureus PBP1.
J.Struct.Biol., 216, 2024
|
|
8H1A
 
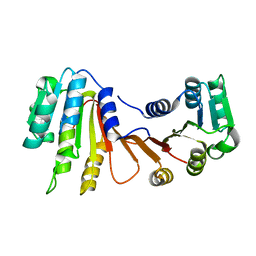 | |
8H27
 
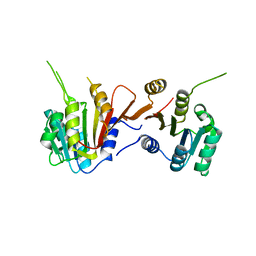 | |
8H0S
 
 | |
8H0T
 
 | |
8H1B
 
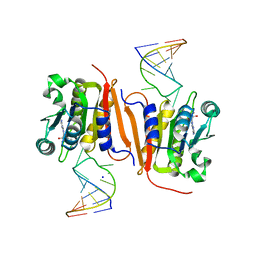 | | Crystal structure of MnmM from S. aureus complexed with SAM and tRNA anti-codon stem loop (ASL) (1.55 A) | | 分子名称: | RNA (5'-R(*AP*CP*GP*GP*AP*CP*UP*UP*UP*GP*AP*CP*UP*CP*CP*GP*U)-3'), S-ADENOSYLMETHIONINE, SODIUM ION, ... | | 著者 | Kim, J, Cho, G, Lee, J. | | 登録日 | 2022-10-01 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Identification of a novel 5-aminomethyl-2-thiouridine methyltransferase in tRNA modification.
Nucleic Acids Res., 51, 2023
|
|
4APP
 
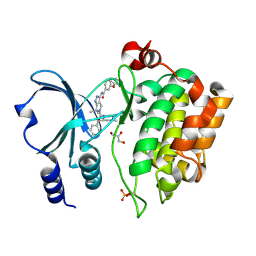 | | Crystal Structure of the Human p21-Activated Kinase 4 in Complex with (S)-N-(5-(3-benzyl-1-methylpiperazine-4-carbonyl)-6,6-dimethyl-1,4,5, 6-tetrahydropyrrolo(3,4-c)pyrazol-3-yl)-3-phenoxybenzamide | | 分子名称: | GLYCEROL, N-[6,6-dimethyl-5-[(2S)-4-methyl-2-(phenylmethyl)piperazin-1-yl]carbonyl-2,4-dihydropyrrolo[3,4-c]pyrazol-3-yl]-3-phenoxy-benzamide, SERINE/THREONINE-PROTEIN KINASE PAK 4 | | 著者 | Knighton, D.D, Deng, Y.L, Wang, C, Guo, C, McAlpine, I, Zhang, J, Kephart, S, Johnson, M.C, Li, H, Bouzida, D, Yang, A, Dong, L, Marakovits, J, Tikhe, J, Richardson, P, Guo, L.C, Kania, R, Edwards, M.P, Kraynov, E, Christensen, J, Piraino, J, Lee, J, Dagostino, E, Del-Carmen, C, Smeal, T, Murray, B.W. | | 登録日 | 2012-04-04 | | 公開日 | 2012-06-06 | | 最終更新日 | 2019-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Discovery of Pyrroloaminopyrazoles as Novel Pak Inhibitors.
J.Med.Chem., 55, 2012
|
|
3TUC
 
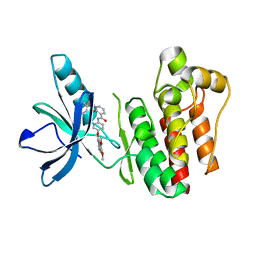 | | Crystal structure of SYK kinase domain with 1-benzyl-N-(5-(6,7-dimethoxyquinolin-4-yloxy)pyridin-2-yl)-2-oxo-1,2-dihydropyridine-3-carboxamide | | 分子名称: | 1-benzyl-N-{5-[(6,7-dimethoxyquinolin-4-yl)oxy]pyridin-2-yl}-2-oxo-1,2-dihydropyridine-3-carboxamide, Tyrosine-protein kinase SYK | | 著者 | Lovering, F, McDonald, J, Whitlock, G, Glossop, P, Phillips, C, Sabnis, Y, Ryan, M, Fitz, L, Lee, J, Chang, J.S, Han, S, Kurumbail, R, Thorarenson, A. | | 登録日 | 2011-09-16 | | 公開日 | 2012-08-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Identification of Type-II Inhibitors Using Kinase Structures.
Chem.Biol.Drug Des., 80, 2012
|
|
3TUD
 
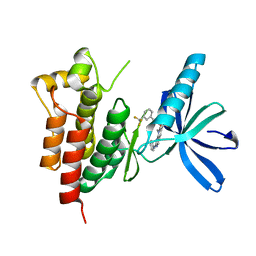 | | Crystal structure of SYK kinase domain with N-(4-methyl-3-(8-methyl-7-oxo-2-(phenylamino)-7,8-dihydropyrido[2,3-d]pyrimidin-6-yl)phenyl)-3-(trifluoromethyl)benzamide | | 分子名称: | N-{4-methyl-3-[8-methyl-7-oxo-2-(phenylamino)-7,8-dihydropyrido[2,3-d]pyrimidin-6-yl]phenyl}-3-(trifluoromethyl)benzamide, Tyrosine-protein kinase SYK | | 著者 | Lovering, F, McDonald, J, Whitlock, G, Glossop, P, Phillips, C, Sabnis, Y, Ryan, M, Fitz, L, Lee, J, Chang, J.S, Han, S, Kurumbail, R, Thorarenson, A. | | 登録日 | 2011-09-16 | | 公開日 | 2012-08-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Identification of Type-II Inhibitors Using Kinase Structures.
Chem.Biol.Drug Des., 80, 2012
|
|
6KZ7
 
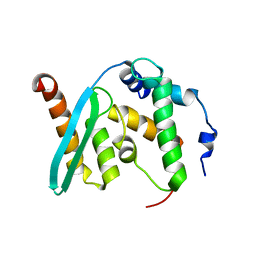 | | The crystal structure of BAF155 SWIRM domain and N-terminal elongated hSNF5 RPT1 domain complex: Chromatin remodeling complex | | 分子名称: | SWI/SNF complex subunit SMARCC1, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | | 著者 | Lee, W, Han, J, Kim, I, Park, J.H, Joo, K, Lee, J, Suh, J.Y. | | 登録日 | 2019-09-23 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | A Coil-to-Helix Transition Serves as a Binding Motif for hSNF5 and BAF155 Interaction.
Int J Mol Sci, 21, 2020
|
|
