8T4E
 
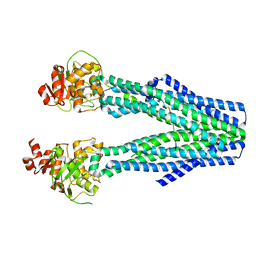 | |
8DRX
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp10-nsp11 (C10) cut site sequence (form 2) | | 分子名称: | Fusion protein of 3C-like proteinase nsp5 and nsp10-nsp11 (C10) cut site, SODIUM ION | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRS
 
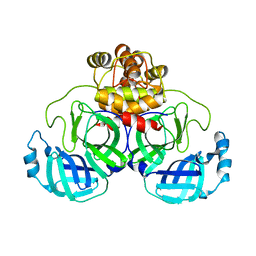 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp6-nsp7 (C6) cut site sequence | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRT
 
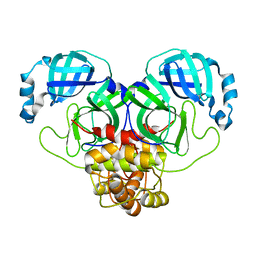 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp6-nsp7 (C6) cut site sequence (form 2) | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRR
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp4-nsp5 (C4) cut site sequence | | 分子名称: | 3C-like proteinase nsp5, SODIUM ION | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRW
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp9-nsp10 (C9) cut site sequence | | 分子名称: | DI(HYDROXYETHYL)ETHER, Fusion protein of 3C-like proteinase nsp5 and nsp9-nsp10 (C9) cut site, PENTAETHYLENE GLYCOL, ... | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRU
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp7-nsp8 (C7) cut site sequence | | 分子名称: | DI(HYDROXYETHYL)ETHER, Fusion protein of 3C-like proteinase nsp5 and nsp7-nsp8 (C7) cut site, PENTAETHYLENE GLYCOL, ... | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRV
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp8-nsp9 (C8) cut site sequence | | 分子名称: | Fusion protein of 3C-like proteinase nsp5 and nsp8-nsp9 (C8) cut site, PENTAETHYLENE GLYCOL | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRY
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp12-nsp13 (C12) cut site sequence | | 分子名称: | DI(HYDROXYETHYL)ETHER, Fusion protein of 3C-like proteinase nsp5 and nsp12-nsp13 (C12) cut site | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DS0
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp14-nsp15 (C14) cut site sequence (form 2) | | 分子名称: | 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DRZ
 
 | | Product structure of SARS-CoV-2 Mpro C145A mutant in complex with nsp13-nsp14 (C13) cut site sequence | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DS2
 
 | | Structure of SARS-CoV-2 Mpro in complex with the nsp13-nsp14 (C13) cut site sequence (form 2) | | 分子名称: | 3C-like proteinase nsp5, GLYCEROL, SODIUM ION | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
8DS1
 
 | | Structure of SARS-CoV-2 Mpro in complex with nsp12-nsp13 (C12) cut site sequence | | 分子名称: | 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER, SODIUM ION | | 著者 | Lee, J, Kenward, C, Worrall, L.J, Vuckovic, M, Paetzel, M, Strynadka, N.C.J. | | 登録日 | 2022-07-21 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation.
Nat Commun, 13, 2022
|
|
2QQR
 
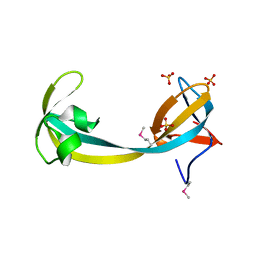 | | JMJD2A hybrid tudor domains | | 分子名称: | JmjC domain-containing histone demethylation protein 3A, SULFATE ION | | 著者 | Lee, J, Botuyan, M.V, Mer, G. | | 登録日 | 2007-07-26 | | 公開日 | 2007-12-11 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor.
Nat.Struct.Mol.Biol., 15, 2008
|
|
2QQS
 
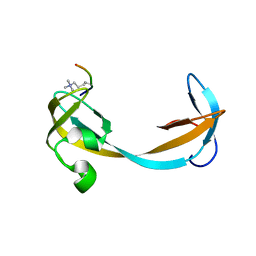 | |
8U29
 
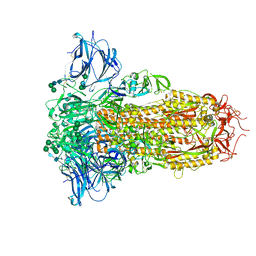 | | Prefusion structure of the PRD-0038 spike glycoprotein ectodomain trimer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PRD-0038 Spike glycoprotein, ... | | 著者 | Lee, J, Park, Y.J, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | 登録日 | 2023-09-05 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Broad receptor tropism and immunogenicity of a clade 3 sarbecovirus.
Cell Host Microbe, 31, 2023
|
|
2VLG
 
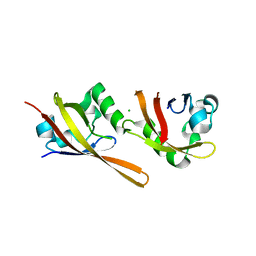 | | KinA PAS-A domain, homodimer | | 分子名称: | ACETATE ION, CHLORIDE ION, SPORULATION KINASE A | | 著者 | Lee, J, Tomchick, D.R, Brautigam, C.A, Machius, M, Kort, R, Hellingwerf, K.J, Gardner, K.H. | | 登録日 | 2008-01-14 | | 公開日 | 2008-03-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Changes at the Kina Pas-A Dimerization Interface Influence Histidine Kinase Function.
Biochemistry, 47, 2008
|
|
2FHD
 
 | | Crystal structure of Crb2 tandem tudor domains | | 分子名称: | DNA repair protein rhp9/CRB2, PHOSPHATE ION | | 著者 | Lee, J, Botuyan, M.V, Thompson, J.R, Mer, G. | | 登録日 | 2005-12-23 | | 公開日 | 2007-01-02 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair.
Cell(Cambridge,Mass.), 127, 2006
|
|
3HL4
 
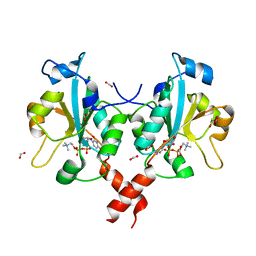 | | Crystal structure of a mammalian CTP:phosphocholine cytidylyltransferase with CDP-choline | | 分子名称: | Choline-phosphate cytidylyltransferase A, FORMIC ACID, GLYCEROL, ... | | 著者 | Lee, J, Paetzel, M, Cornell, R.B. | | 登録日 | 2009-05-26 | | 公開日 | 2009-09-22 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of a mammalian CTP: Phosphocholine cytidylyltransferase catalytic domain reveals novel active site residues within a highly conserved nucleotidyl-transferase fold
J.Biol.Chem., 284, 2009
|
|
2AQZ
 
 | | Crystal structure of FGF-1, S17T/N18T/G19 deletion mutant | | 分子名称: | Heparin-binding growth factor 1, SULFATE ION | | 著者 | Lee, J, Blaber, M. | | 登録日 | 2005-08-18 | | 公開日 | 2006-02-07 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Conversion of type I 4:6 to 3:5 beta-turn types in human acidic fibroblast growth factor: Effects upon structure, stability, folding, and mitogenic function.
Proteins, 62, 2006
|
|
2B02
 
 | | Crystal Structure of ARNT PAS-B Domain | | 分子名称: | Aryl hydrocarbon receptor nuclear translocator | | 著者 | Lee, J, Botuyan, M.V, Nomine, Y, Ohh, M, Thompson, J.R, Mer, G. | | 登録日 | 2005-09-12 | | 公開日 | 2006-10-24 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal Structure and Binding Properties of ARNT PAS-B Heterodimerization Domain
To be Published
|
|
1Z2V
 
 | |
1Z4S
 
 | |
1YTO
 
 | | Crystal Structure of Gly19 deletion Mutant of Human Acidic Fibroblast Growth Factor | | 分子名称: | FORMIC ACID, Heparin-binding growth factor 1, SULFATE ION | | 著者 | Lee, J, Blaber, M. | | 登録日 | 2005-02-10 | | 公開日 | 2006-01-24 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Conversion of type I 4:6 to 3:5 beta-turn types in human acidic fibroblast growth factor: Effects upon structure, stability, folding, and mitogenic function.
Proteins, 62, 2006
|
|
3EQA
 
 | | Catalytic domain of glucoamylase from aspergillus niger complexed with tris and glycerol | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Glucoamylase, ... | | 著者 | Lee, J, Paetzel, M. | | 登録日 | 2008-09-30 | | 公開日 | 2009-10-13 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of the catalytic domain of glucoamylase from Aspergillus niger.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
