7JP1
 
 | |
7KHP
 
 | | Acyl-enzyme intermediate structure of SARS-CoV-2 Mpro in complex with its C-terminal autoprocessing sequence. | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Lee, J, Worrall, L.J, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2020-10-21 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallographic structure of wild-type SARS-CoV-2 main protease acyl-enzyme intermediate with physiological C-terminal autoprocessing site.
Nat Commun, 11, 2020
|
|
7DOG
 
 | |
6IKX
 
 | |
6IKZ
 
 | |
7K2V
 
 | |
7K1W
 
 | |
7K1Y
 
 | |
6K8D
 
 | |
6KNL
 
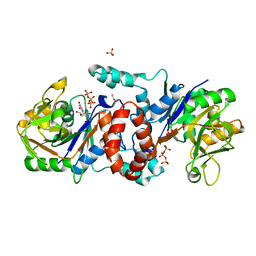 | |
6KNJ
 
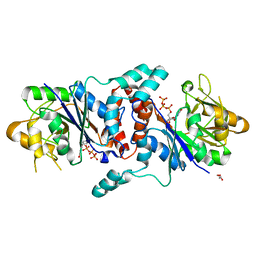 | |
4V46
 
 | | Crystal structure of the BAFF-BAFF-R complex | | Descriptor: | MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 13B, Tumor necrosis factor receptor superfamily member 13C | | Authors: | Kim, H.M, Yu, K.S, Lee, M.E, Shin, D.R, Kim, Y.S, Paik, S.G, Yoo, O.J, Lee, H, Lee, J.-O. | | Deposit date: | 2003-03-23 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the BAFF-BAFF-R complex and its implications for receptor activation
NAT.STRUCT.BIOL., 10, 2003
|
|
1EL4
 
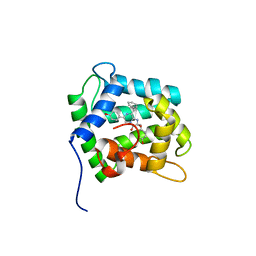 | | STRUCTURE OF THE CALCIUM-REGULATED PHOTOPROTEIN OBELIN DETERMINED BY SULFUR SAS | | Descriptor: | C2-HYDROXY-COELENTERAZINE, CHLORIDE ION, OBELIN | | Authors: | Liu, Z.J, Vysotski, E.S, Rose, J, Lee, J, Wang, B.C. | | Deposit date: | 2000-03-13 | | Release date: | 2001-03-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure of the Ca2+-regulated photoprotein obelin at 1.7 A resolution determined directly from its sulfur substructure.
Protein Sci., 9, 2000
|
|
8VBW
 
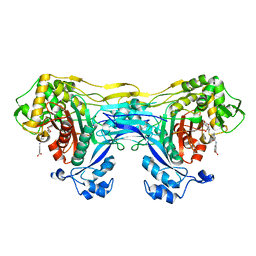 | | Structure of the monofunctional Staphylococcus aureus PBP1 in its beta-lactam (Ertapenem) inhibited form | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein 1 | | Authors: | Bon, C.G, Lee, J, Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic analysis of the monofunctional Staphylococcus aureus PBP1.
J.Struct.Biol., 216, 2024
|
|
8VBT
 
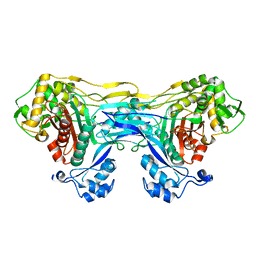 | |
8VBU
 
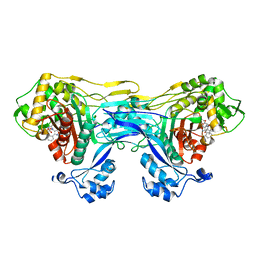 | | Structure of the monofunctional Staphylococcus aureus PBP1 in its beta-lactam (Oxacillin) inhibited form | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, Penicillin-binding protein 1 | | Authors: | Bon, C.G, Lee, J, Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and kinetic analysis of the monofunctional Staphylococcus aureus PBP1.
J.Struct.Biol., 216, 2024
|
|
8VBV
 
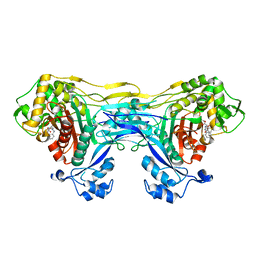 | | Structure of the monofunctional Staphylococcus aureus PBP1 in its beta-lactam (Cephalexin) inhibited form | | Descriptor: | (2S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5-methyl-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Penicillin-binding protein 1 | | Authors: | Bon, C.G, Lee, J, Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and kinetic analysis of the monofunctional Staphylococcus aureus PBP1.
J.Struct.Biol., 216, 2024
|
|
4UIP
 
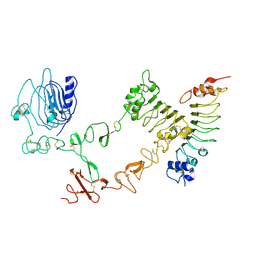 | | The complex structure of extracellular domain of EGFR with Repebody (rAC1). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, EPIDERMAL GROWTH FACTOR RECEPTOR, ... | | Authors: | Kang, Y.J, Cha, Y.J, Cho, H.S, Lee, J.J, Kim, H.S. | | Deposit date: | 2015-03-31 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Enzymatic Prenylation and Oxime Ligation for the Synthesis of Stable and Homogeneous Protein-Drug Conjugates for Targeted Therapy.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4XSJ
 
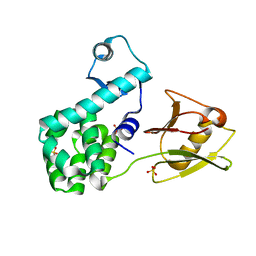 | | Crystal structure of the N-terminal domain of the human mitochondrial calcium uniporter fused with T4 lysozyme | | Descriptor: | Lysozyme,Calcium uniporter protein, mitochondrial, SULFATE ION | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-01-22 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
4XTB
 
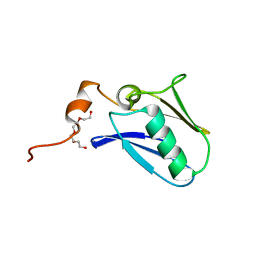 | | Crystal structure of the N-terminal domain of the human mitochondrial calcium uniporter | | Descriptor: | Calcium uniporter protein, mitochondrial, TETRAETHYLENE GLYCOL | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-01-23 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
7CM4
 
 | | Crystal Structure of COVID-19 virus spike receptor-binding domain complexed with a neutralizing antibody CT-P59 | | Descriptor: | 1,2-ETHANEDIOL, IgG heavy chain, IgG light chain, ... | | Authors: | Kim, Y.G, Jeong, J.H, Bae, J.S, Lee, J. | | Deposit date: | 2020-07-24 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein.
Nat Commun, 12, 2021
|
|
8CUB
 
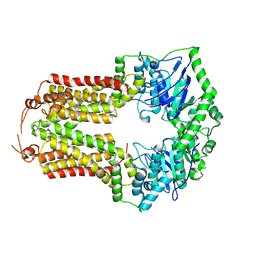 | |
1C02
 
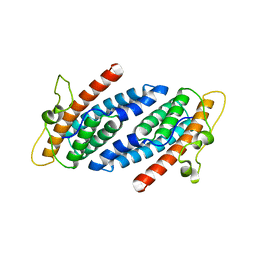 | | CRYSTAL STRUCTURE OF YEAST YPD1P | | Descriptor: | PHOSPHOTRANSFERASE YPD1P | | Authors: | Song, H.K, Lee, J.Y, Lee, M.G, Suh, S.W. | | Deposit date: | 1999-07-14 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into eukaryotic multistep phosphorelay signal transduction revealed by the crystal structure of Ypd1p from Saccharomyces cerevisiae.
J.Mol.Biol., 293, 1999
|
|
1C03
 
 | | CRYSTAL STRUCTURE OF YPD1P (TRICLINIC FORM) | | Descriptor: | HYPOTHETICAL PROTEIN YDL235C | | Authors: | Song, H.K, Lee, J.Y, Lee, M.G, Suh, S.W. | | Deposit date: | 1999-07-14 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into eukaryotic multistep phosphorelay signal transduction revealed by the crystal structure of Ypd1p from Saccharomyces cerevisiae.
J.Mol.Biol., 293, 1999
|
|
8HGU
 
 | |
