7WVF
 
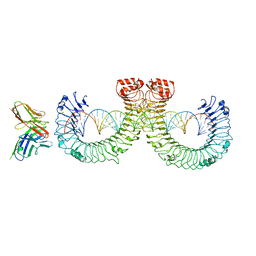 | | ectoTLR3-mAb12-poly(I:C) complex | | Descriptor: | RNA (46-MER), Toll-like receptor 3, mAb12 | | Authors: | Lim, C.S, Jang, Y.H, Lee, G.Y, Han, G.M, Lee, J.O. | | Deposit date: | 2022-02-10 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | TLR3 forms a highly organized cluster when bound to a poly(I:C) RNA ligand.
Nat Commun, 13, 2022
|
|
7WVJ
 
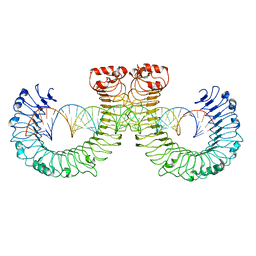 | | NT-mut(K117D,K139D,K145D) TLR3 -poly I:C complex | | Descriptor: | RNA (46-MER), Toll-like receptor 3 | | Authors: | Lim, C.S, Jang, Y.H, Lee, G.Y, Han, G.M, Lee, J.O. | | Deposit date: | 2022-02-10 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | TLR3 forms a highly organized cluster when bound to a poly(I:C) RNA ligand.
Nat Commun, 13, 2022
|
|
7WV4
 
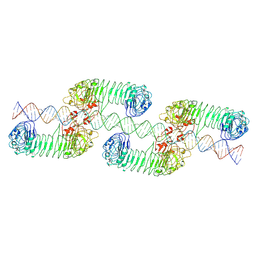 | | ectoTLR3-poly(I:C) cluster | | Descriptor: | RNA (80-MER), Toll-like receptor 3 | | Authors: | Lim, C.S, Jang, Y.H, Lee, G.Y, Han, G.M, Lee, J.O. | | Deposit date: | 2022-02-09 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | TLR3 forms a highly organized cluster when bound to a poly(I:C) RNA ligand.
Nat Commun, 13, 2022
|
|
7WVE
 
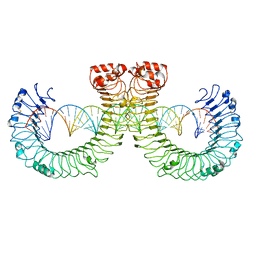 | | CT-mut (D523K,D524K,E527K) TLR3-poly(I:C) complex | | Descriptor: | RNA (46-MER), Toll-like receptor 3 | | Authors: | Lim, C.S, Jang, Y.H, Lee, G.Y, Han, G.M, Lee, J.O. | | Deposit date: | 2022-02-10 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | TLR3 forms a highly organized cluster when bound to a poly(I:C) RNA ligand.
Nat Commun, 13, 2022
|
|
2A8D
 
 | | Haemophilus influenzae beta-carbonic anhydrase complexed with bicarbonate | | Descriptor: | BICARBONATE ION, Carbonic anhydrase 2, SULFATE ION, ... | | Authors: | Cronk, J.D, Rowlett, R.S, Zhang, K.Y.J, Tu, C, Endrizzi, J.A, Lee, J, Gareiss, P.C, Preiss, J.R. | | Deposit date: | 2005-07-07 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of a Novel Noncatalytic Bicarbonate Binding Site in Eubacterial beta-Carbonic Anhydrase
Biochemistry, 45, 2006
|
|
2A8C
 
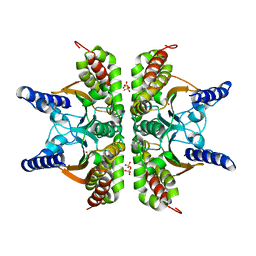 | | Haemophilus influenzae beta-carbonic anhydrase | | Descriptor: | Carbonic anhydrase 2, SULFATE ION, ZINC ION | | Authors: | Cronk, J.D, Rowlett, R.S, Zhang, K.Y.J, Tu, C, Endrizzi, J.A, Lee, J, Gareiss, P.C, Preiss, J.R. | | Deposit date: | 2005-07-07 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of a Novel Noncatalytic Bicarbonate Binding Site in Eubacterial beta-Carbonic Anhydrase
Biochemistry, 45, 2006
|
|
2AEP
 
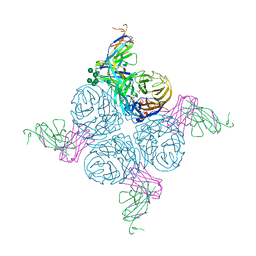 | | An epidemiologically significant epitope of a 1998 influenza virus neuraminidase forms a highly hydrated interface in the NA-antibody complex. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, FAB heavy chain, ... | | Authors: | Venkatramani, L, Bochkareva, E, Lee, J.T, Gulati, U, Laver, W.G, Bochkarev, A, Air, G.M. | | Deposit date: | 2005-07-23 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An Epidemiologically Significant Epitope of a 1998 Human Influenza Virus Neuraminidase Forms a Highly Hydrated Interface in the NA-Antibody Complex
J.Mol.Biol., 356, 2006
|
|
7X89
 
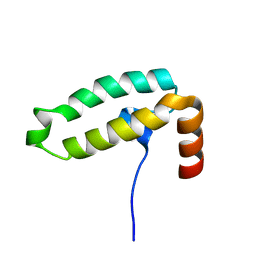 | | Tid1 | | Descriptor: | DnaJ homolog subfamily A member 3, mitochondrial | | Authors: | Jang, J, Lee, S.H, Kang, D.H, Sim, D.W, Jo, K.S, Ryu, H, Kim, E.H, Ryu, K.S, Lee, J.H, Kim, J.H, Won, H.S. | | Deposit date: | 2022-03-11 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural studies on the J-domain and GF-motif of the mitochondrial Hsp40, Tid1
To Be Published
|
|
2AZ2
 
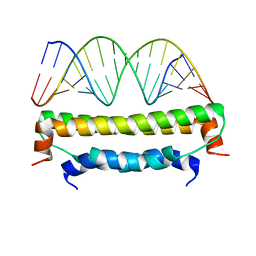 | | Flock House virus B2-dsRNA Complex (P4122) | | Descriptor: | 5'-R(*GP*CP*AP*(5BU)P*GP*GP*AP*CP*GP*CP*GP*(5BU)P*CP*CP*AP*(5BU)P*GP*C)-3', B2 protein | | Authors: | Chao, J.A, Lee, J.H, Chapados, B.R, Debler, E.W, Schneemann, A, Williamson, J.R. | | Deposit date: | 2005-09-09 | | Release date: | 2005-10-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dual modes of RNA-silencing suppression by Flock House virus protein B2.
Nat.Struct.Mol.Biol., 12, 2005
|
|
4M4D
 
 | | Crystal structure of lipopolysaccharide binding protein | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipopolysaccharide-binding protein | | Authors: | Eckert, J.K, Kim, Y.J, Kim, J.I, Gurtler, K, Oh, D.Y, Ploeg, A.H, Pickkers, P, Lundvall, L, Hamann, L, Giamarellos-Bourboulis, E, Kubarenko, A.V, Weber, A.N, Kabesch, M, Kumpf, O, An, H.J, Lee, J.O, Schumann, R.R. | | Deposit date: | 2013-08-07 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.909 Å) | | Cite: | The crystal structure of lipopolysaccharide binding protein reveals the location of a frequent mutation that impairs innate immunity.
Immunity, 39, 2013
|
|
3BL6
 
 | | Crystal structure of Staphylococcus aureus 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase in complex with formycin A | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, 5'-methylthioadenosine nucleosidase/S-adenosylhomocysteine nucleosidase | | Authors: | Siu, K.K.W, Lee, J.E, Smith, G.D, Horvatin, C, Howell, P.L. | | Deposit date: | 2007-12-10 | | Release date: | 2008-06-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Staphylococcus aureus 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2AGG
 
 | | succinyl-AAPK-trypsin acyl-enzyme at 1.28 A resolution | | Descriptor: | CALCIUM ION, Cationic trypsin, SULFATE ION, ... | | Authors: | Radisky, E.S, Lee, J.M, Lu, C.J, Koshland Jr, D.E. | | Deposit date: | 2005-07-26 | | Release date: | 2006-05-16 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Insights into the serine protease mechanism from atomic resolution structures of trypsin reaction intermediates
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
4MN0
 
 | | Spatial structure of the novel light-sensitive photoprotein berovin from the ctenophore Beroe abyssicola in the Ca2+-loaded apoprotein conformation state | | Descriptor: | Berovin, CALCIUM ION, MAGNESIUM ION | | Authors: | Liu, Z.J, Stepanyuk, G.A, Vysotski, E.S, Lee, J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2013-09-09 | | Release date: | 2013-10-16 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Spatial structure of the novel light-sensitive photoprotein berovin from the ctenophore Beroe abyssicola in the Ca(2+)-loaded apoprotein conformation state.
Biochim.Biophys.Acta, 1834, 2013
|
|
4N1G
 
 | | Crystal Structure of Ca(2+)- discharged F88Y obelin mutant from Obelia longissima at 1.50 Angstrom resolution | | Descriptor: | CALCIUM ION, N-[3-BENZYL-5-(4-HYDROXYPHENYL)PYRAZIN-2-YL]-2-(4-HYDROXYPHENYL)ACETAMIDE, Obelin | | Authors: | Natashin, P.V, Markova, S.V, Lee, J, Vysotski, E.S, Liu, Z.J. | | Deposit date: | 2013-10-04 | | Release date: | 2014-02-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of the F88Y obelin mutant before and after bioluminescence provide molecular insight into spectral tuning among hydromedusan photoproteins
Febs J., 281, 2014
|
|
8IS3
 
 | | Structural model for the micelle-bound indolicidin-like peptide in solution | | Descriptor: | Indolicidin-like antimicrobial peptide | | Authors: | Kim, B, Ko, Y.H, Kim, J, Lee, J, Nam, C.H, Kim, J.H. | | Deposit date: | 2023-03-20 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structural model for the micelle-bound indolicidin-like peptide in solution
To Be Published
|
|
5WUV
 
 | | Crystal structure of Certolizumab Fab | | Descriptor: | heavy chain, light chain | | Authors: | Heo, Y.S, Lee, J.U, Son, J.Y, Shin, W, Yoo, K.Y. | | Deposit date: | 2016-12-21 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Molecular Basis for the Neutralization of Tumor Necrosis Factor alpha by Certolizumab Pegol in the Treatment of Inflammatory Autoimmune Diseases
Int J Mol Sci, 18, 2017
|
|
4MRY
 
 | | Crystal Structure of Ca(2+)- discharged Y138F obelin mutant from Obelia longissima at 1.30 Angstrom resolution | | Descriptor: | CALCIUM ION, N-[3-BENZYL-5-(4-HYDROXYPHENYL)PYRAZIN-2-YL]-2-(4-HYDROXYPHENYL)ACETAMIDE, Obelin | | Authors: | Natashin, P.V, Ding, W, Eremeeva, E.V, Markova, S.V, Lee, J, Vysotski, E.S, Liu, Z.J. | | Deposit date: | 2013-09-17 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.299 Å) | | Cite: | Structures of the Ca2+-regulated photoprotein obelin Y138F mutant before and after bioluminescence support the catalytic function of a water molecule in the reaction.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8ISO
 
 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Beta-lactamase | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
8ISR
 
 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by cefaclor | | Descriptor: | (R)-2-((R)-((R)-2-amino-2-phenylacetamido)(carboxy)methyl)-5-chloro-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
8ISP
 
 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by cephalexin | | Descriptor: | (R)-2-((R)-((R)-2-amino-2-phenylacetamido)(carboxy)methyl)-5-methyl-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
8ISQ
 
 | | Crystal structure of extended-spectrum class A beta-lactamase, CESS-1 E166Q acylated by ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, ... | | Authors: | Jeong, B.G, Kim, M.Y, Jeong, C.S, Do, H.W, Lee, J.H, Cha, S.S. | | Deposit date: | 2023-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Characterization of the extended substrate spectrum of the class A beta-lactamase CESS-1 from Stenotrophomonas sp. and structure-based investigation into its substrate preference.
Int J Antimicrob Agents, 63, 2024
|
|
5W8N
 
 | |
5W8X
 
 | | Lipid A Disaccharide Synthase (LpxB)-7 solubilizing mutations-Bound to UDP | | Descriptor: | Lipid-A-disaccharide synthase, URIDINE-5'-DIPHOSPHATE | | Authors: | Bohl, T.E, Aihara, H, Shi, K, Lee, J.K. | | Deposit date: | 2017-06-22 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structure of lipid A disaccharide synthase LpxB from Escherichia coli.
Nat Commun, 9, 2018
|
|
5W8S
 
 | | Lipid A Disaccharide Synthase (LpxB)-7 solubilizing mutations | | Descriptor: | LITHIUM ION, Lipid-A-disaccharide synthase, SODIUM ION | | Authors: | Bohl, T.E, Aihara, H, Shi, K, Lee, J.K. | | Deposit date: | 2017-06-22 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of lipid A disaccharide synthase LpxB from Escherichia coli.
Nat Commun, 9, 2018
|
|
7C04
 
 | | Crystal structure of human Trap1 with DN203492 | | Descriptor: | 4-chloranyl-1-[[2-methoxy-4-(trifluoromethyl)phenyl]methyl]pyrazolo[3,4-d]pyrimidin-6-amine, Heat shock protein 75 kDa, mitochondrial | | Authors: | Kim, D, Kim, D, Kim, S.Y, Lee, J.H, Kang, B.H, Kang, S, Lee, C. | | Deposit date: | 2020-04-30 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of pyrazolo[3,4-d]pyrimidine-6-amine-based TRAP1 inhibitors that demonstrate in vivo anticancer activity in mouse xenograft models.
Bioorg.Chem., 101, 2020
|
|
