7DHA
 
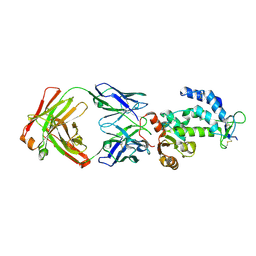 | | crystal structure of CD38 in complex with daratumumab | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, Heavy Chain, Light chain | | Authors: | Lee, H.T, Heo, Y.S. | | Deposit date: | 2020-11-13 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of CD38 in complex with daratumumab, a first-in-class anti-CD38 antibody drug for treating multiple myeloma.
Biochem.Biophys.Res.Commun., 536, 2021
|
|
7EOW
 
 | |
3JBJ
 
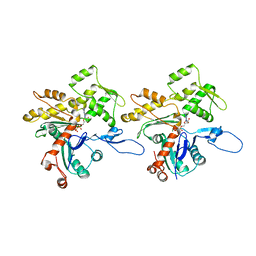 | | Cryo-EM reconstruction of F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Kim, L.Y, Thompson, P.M, Lee, H.T, Pershad, M, Campbell, S.L, Alushin, G.M. | | Deposit date: | 2015-09-03 | | Release date: | 2015-11-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | The Structural Basis of Actin Organization by Vinculin and Metavinculin.
J.Mol.Biol., 428, 2016
|
|
5X8M
 
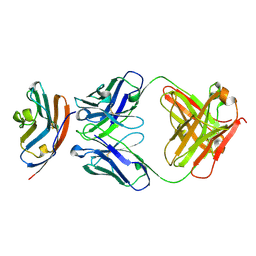 | | PD-L1 in complex with durvalumab | | Descriptor: | Programmed cell death 1 ligand 1, durvalumab heavy chain, durvalumab light chain | | Authors: | Heo, Y.S, Lee, H.T. | | Deposit date: | 2017-03-03 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.661 Å) | | Cite: | Molecular mechanism of PD-1/PD-L1 blockade via anti-PD-L1 antibodies atezolizumab and durvalumab
Sci Rep, 7, 2017
|
|
5X8L
 
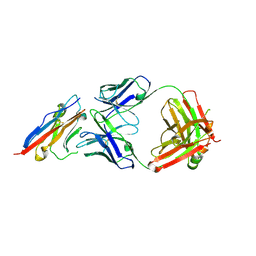 | | PD-L1 in complex with atezolizumab | | Descriptor: | Programmed cell death 1 ligand 1, atezolizumab heavy chain, atezolizumab light chain | | Authors: | Heo, Y.S, Lee, H.T. | | Deposit date: | 2017-03-03 | | Release date: | 2017-08-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular mechanism of PD-1/PD-L1 blockade via anti-PD-L1 antibodies atezolizumab and durvalumab
Sci Rep, 7, 2017
|
|
1ZOI
 
 | | Crystal Structure of a Stereoselective Esterase from Pseudomonas putida IFO12996 | | Descriptor: | esterase | | Authors: | Elmi, F, Lee, H.T, Huang, J.Y, Hsieh, Y.C, Wang, Y.L, Chen, Y.J, Shaw, S.Y, Chen, C.J. | | Deposit date: | 2005-05-13 | | Release date: | 2006-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Stereoselective esterase from Pseudomonas putida IFO12996 reveals alpha/beta hydrolase folds for D-beta-acetylthioisobutyric acid synthesis
J.Bacteriol., 187, 2005
|
|
7BXA
 
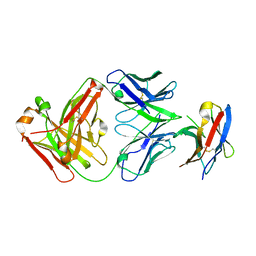 | | Crystal structure of PD-1 in complex with tislelizumab Fab | | Descriptor: | Programmed cell death protein 1, heavy chain, light chain | | Authors: | Heo, Y.S, Lee, S.H, Lim, H, Lee, H.T, Kim, Y.J, Park, E.B. | | Deposit date: | 2020-04-18 | | Release date: | 2020-06-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Crystal structure of PD-1 in complex with an antibody-drug tislelizumab used in tumor immune checkpoint therapy.
Biochem.Biophys.Res.Commun., 527, 2020
|
|
3JBK
 
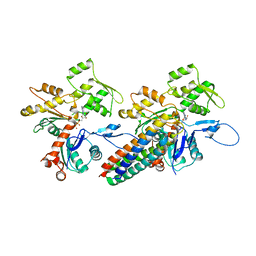 | | Cryo-EM reconstruction of the metavinculin-actin interface | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Kim, L.Y, Thompson, P.M, Lee, H.T, Pershad, M, Campbell, S.L, Alushin, G.M. | | Deposit date: | 2015-09-03 | | Release date: | 2015-11-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | The Structural Basis of Actin Organization by Vinculin and Metavinculin.
J.Mol.Biol., 428, 2016
|
|
3JBI
 
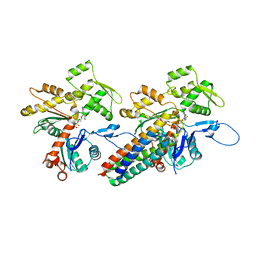 | | MDFF model of the vinculin tail domain bound to F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Kim, L.Y, Thompson, P.M, Lee, H.T, Pershad, M, Campbell, S.L, Alushin, G.M. | | Deposit date: | 2015-09-02 | | Release date: | 2015-11-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | The Structural Basis of Actin Organization by Vinculin and Metavinculin.
J.Mol.Biol., 428, 2016
|
|
7WWP
 
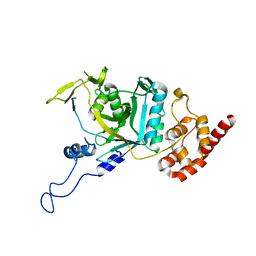 | | Crystal structure of human Npl4 | | Descriptor: | Nuclear protein localization protein 4 homolog, ZINC ION | | Authors: | Nguyen, T.Q, Le, L.T.M, Kim, D.H, Ko, K.S, Lee, H.T, Nguyen, Y.T.K, Kim, H.S, Han, B.W, Kang, W, Yang, J.K. | | Deposit date: | 2022-02-14 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis for the interaction between human Npl4 and Npl4-binding motif of human Ufd1.
Structure, 30, 2022
|
|
7WWQ
 
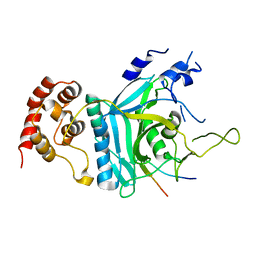 | | Crystal structure of human Ufd1-Npl4 complex | | Descriptor: | Nuclear protein localization protein 4 homolog, Ubiquitin recognition factor in ER-associated degradation protein 1 | | Authors: | Nguyen, T.Q, Le, L.T.M, Kim, D.H, Ko, K.S, Lee, H.T, Nguyen, Y.T.K, Kim, H.S, Han, B.W, Kang, W, Yang, J.K. | | Deposit date: | 2022-02-14 | | Release date: | 2022-09-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural basis for the interaction between human Npl4 and Npl4-binding motif of human Ufd1.
Structure, 30, 2022
|
|
