6LRG
 
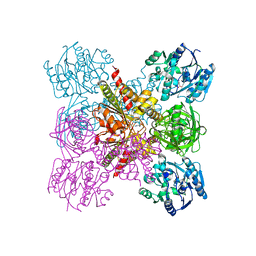 | | Crystal Structure of the Ternary Complex of AgrE with Ornithine and NAD+ | | Descriptor: | Alr4995 protein, L-ornithine, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lee, H, Rhee, S. | | Deposit date: | 2020-01-16 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.41218114 Å) | | Cite: | Structural and mutational analyses of the bifunctional arginine dihydrolase and ornithine cyclodeaminase AgrE from the cyanobacteriumAnabaena.
J.Biol.Chem., 295, 2020
|
|
6UH7
 
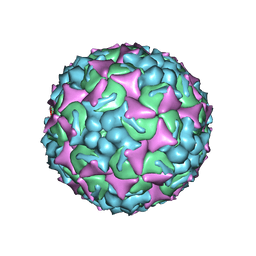 | | EV-A71 strain 11316 complexed with MADAL compound 30 | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Lee, H, Hafenstein, S. | | Deposit date: | 2019-09-26 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Scaffold Simplification Strategy Leads to a Novel Generation of Dual Human Immunodeficiency Virus and Enterovirus-A71 Entry Inhibitors.
J.Med.Chem., 63, 2020
|
|
6UH6
 
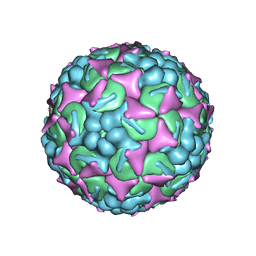 | | EV-A71 strain 11316 complexed with MADAL compound 22 | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Lee, H, Hafenstein, S. | | Deposit date: | 2019-09-26 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Scaffold Simplification Strategy Leads to a Novel Generation of Dual Human Immunodeficiency Virus and Enterovirus-A71 Entry Inhibitors.
J.Med.Chem., 63, 2020
|
|
6UH1
 
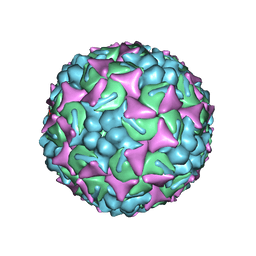 | | Structure of the EVA71 strain 11316 capsid | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Lee, H, Hafenstein, S. | | Deposit date: | 2019-09-26 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Scaffold Simplification Strategy Leads to a Novel Generation of Dual Human Immunodeficiency Virus and Enterovirus-A71 Entry Inhibitors.
J.Med.Chem., 63, 2020
|
|
3J3Z
 
 | | Structure of MA28-7 neutralizing antibody Fab fragment from electron cryo-microscopy of enterovirus 71 complexed with a Fab fragment | | Descriptor: | MA28-7 neutralizing antibody heavy chain, MA28-7 neutralizing antibody light chain | | Authors: | Lee, H, Cifuente, J.O, Ashley, R.E, Conway, J.F, Makhov, A.M, Tano, Y, Shimizu, H, Nishimura, Y, Hafenstein, S. | | Deposit date: | 2013-05-21 | | Release date: | 2013-08-28 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (23.4 Å) | | Cite: | A strain-specific epitope of enterovirus 71 identified by cryo-electron microscopy of the complex with fab from neutralizing antibody.
J.Virol., 87, 2013
|
|
6OAS
 
 | |
5XQH
 
 | | Crystal structure of truncated human Rogdi | | Descriptor: | Protein rogdi homolog | | Authors: | Lee, H, Lee, C. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The crystal structure of human Rogdi provides insight into the causes of Kohlschutter-Tonz Syndrome
Sci Rep, 7, 2017
|
|
5XQI
 
 | | Crystal structure of full-length human Rogdi | | Descriptor: | Protein rogdi homolog | | Authors: | Lee, H, Lee, C. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of human Rogdi provides insight into the causes of Kohlschutter-Tonz Syndrome
Sci Rep, 7, 2017
|
|
6LRH
 
 | |
6LRF
 
 | | Crystal structure of unliganded AgrE | | Descriptor: | Alr4995 protein | | Authors: | Lee, H, Rhee, S. | | Deposit date: | 2020-01-16 | | Release date: | 2020-04-01 | | Last modified: | 2020-05-13 | | Method: | X-RAY DIFFRACTION (2.05466056 Å) | | Cite: | Structural and mutational analyses of the bifunctional arginine dihydrolase and ornithine cyclodeaminase AgrE from the cyanobacteriumAnabaena.
J.Biol.Chem., 295, 2020
|
|
7XW3
 
 | |
7XW2
 
 | |
5Y2Y
 
 | | Crystal structure of HaloTag (M175C) complexed with dansyl-PEG2-HaloTag ligand | | Descriptor: | 5-(dimethylamino)-~{N}-[2-(2-hexoxyethoxy)ethyl]naphthalene-1-sulfonamide, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lee, H, Kang, M, Rhee, H, Lee, C. | | Deposit date: | 2017-07-27 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure-guided synthesis of a protein-based fluorescent sensor for alkyl halides
Chem. Commun. (Camb.), 53, 2017
|
|
5Y2X
 
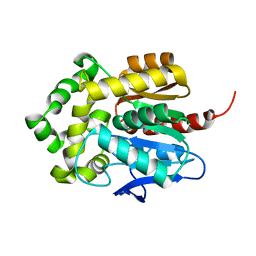 | | Crystal structure of apo-HaloTag (M175C) | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lee, H, Kang, M, Rhee, H, Lee, C. | | Deposit date: | 2017-07-27 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-guided synthesis of a protein-based fluorescent sensor for alkyl halides
Chem. Commun. (Camb.), 53, 2017
|
|
3JD7
 
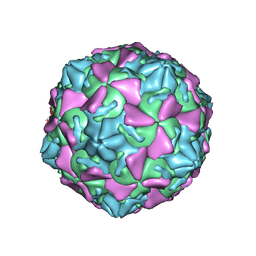 | | The novel asymmetric entry intermediate of a picornavirus captured with nanodiscs | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Lee, H, Shingler, K.L, Organtini, L.J, Ashley, R.E, Makhov, A.M, Conway, J.F, Hafenstein, S. | | Deposit date: | 2016-04-29 | | Release date: | 2016-09-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The novel asymmetric entry intermediate of a picornavirus captured with nanodiscs.
Sci Adv, 2, 2016
|
|
3J7E
 
 | | Electron cryo-microscopy of human papillomavirus 16 and H16.V5 Fab fragments | | Descriptor: | H16.V5 Fab heavy chain, H16.V5 Fab light chain | | Authors: | Lee, H, Brendle, S.A, Bywaters, S.M, Christensen, N.D, Hafenstein, S. | | Deposit date: | 2014-06-23 | | Release date: | 2014-11-26 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (13.6 Å) | | Cite: | A cryo-electron microscopy study identifies the complete H16.V5 epitope and reveals global conformational changes initiated by binding of the neutralizing antibody fragment.
J.Virol., 89, 2015
|
|
3J7G
 
 | |
5JWT
 
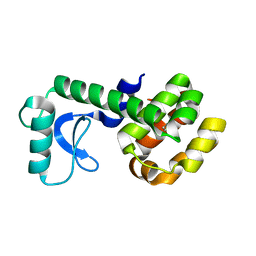 | | T4 Lysozyme L99A/M102Q with Benzene Bound | | Descriptor: | BENZENE, Endolysin | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
5JWU
 
 | | T4 Lysozyme L99A/M102Q with 1,2-Dihydro-1,2-azaborine Bound | | Descriptor: | 1,2-dihydro-1,2-azaborinine, CHLORIDE ION, Endolysin | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
5JWS
 
 | | T4 Lysozyme L99A with 1-Hydro-2-ethyl-1,2-azaborine Bound | | Descriptor: | 2-ethyl-1,2-dihydro-1,2-azaborinine, CHLORIDE ION, Endolysin | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
5JWW
 
 | | T4 Lysozyme L99A/M102Q with 1-Hydro-2-ethyl-1,2-azaborine Bound | | Descriptor: | 2-ethyl-1,2-dihydro-1,2-azaborinine, CHLORIDE ION, Endolysin | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
5JWV
 
 | | T4 Lysozyme L99A/M102Q with Ethylbenzene Bound | | Descriptor: | Endolysin, PHENYLETHANE | | Authors: | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | Deposit date: | 2016-05-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
2LRQ
 
 | | Chemical Shift Assignment and Solution Structure of Fr822A from Drosophila melanogaster. Northeast Structural Genomics Consortium Target Fr822A | | Descriptor: | NuA4 complex subunit EAF3 homolog | | Authors: | Lee, H, Lee, D, Kohan, E, Janjua, H, Xiao, R, Acton, T, Everett, J.K, Montelione, G, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG), Chaperone-Enabled Studies of Epigenetic Regulation Enzymes (CEBS) | | Deposit date: | 2012-04-11 | | Release date: | 2012-07-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Fr822A from Drosophila melanogaster.
To be Published
|
|
7B8W
 
 | | Structure of LIMK1 Kinase domain with allosteric inhibitor TH-470 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-methylpropanoylamino)-~{N}-[2-[(phenylmethyl)-[4-(phenylsulfamoyl)phenyl]carbonyl-amino]ethyl]-1,3-thiazole-5-carboxamide, LIM domain kinase 1 | | Authors: | Lee, H, Yosaatmadja, Y, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Elkins, J.M. | | Deposit date: | 2020-12-13 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of LIMK1 Kinase domain with allosteric inhibitor TH-470
To Be Published
|
|
7CBC
 
 | |
