4W8C
 
 | |
6CSV
 
 | | The structure of the Cep63-Cep152 heterotetrameric complex | | Descriptor: | Centrosomal protein of 63 kDa,Centrosomal protein of 152 kDa | | Authors: | Lee, E, Chen, Y, Zhang, L, Kim, T.S, Ahn, J.I, Park, J.E, Lee, K.S. | | Deposit date: | 2018-03-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular architecture of a cylindrical self-assembly at human centrosomes.
Nat Commun, 10, 2019
|
|
6CSU
 
 | | The structure of the Cep63-Cep152 heterotetrameric complex | | Descriptor: | Centrosomal protein of 152 kDa, Centrosomal protein of 63 kDa | | Authors: | Lee, E, Chen, Y, Zhang, L, Kim, T.S, Ahn, J.I, Park, J.E, Lee, K.S. | | Deposit date: | 2018-03-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular architecture of a cylindrical self-assembly at human centrosomes.
Nat Commun, 10, 2019
|
|
4TR1
 
 | | Crystal structure of GSH-bound cGrx2/C15S | | Descriptor: | GLUTATHIONE, Glutaredoxin 3 | | Authors: | Lee, E.H, Hwang, K.Y. | | Deposit date: | 2014-06-13 | | Release date: | 2014-10-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.582 Å) | | Cite: | The GSH- and GSSG-bound structures of glutaredoxin from Clostridium oremlandii.
Arch.Biochem.Biophys., 564C, 2014
|
|
4TR0
 
 | | Crystal structure of GSSG-bound cGrx2 | | Descriptor: | ACETATE ION, Glutaredoxin 3, OXIDIZED GLUTATHIONE DISULFIDE | | Authors: | Lee, E.H, Hwang, K.Y. | | Deposit date: | 2014-06-13 | | Release date: | 2014-10-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | The GSH- and GSSG-bound structures of glutaredoxin from Clostridium oremlandii.
Arch.Biochem.Biophys., 564C, 2014
|
|
2YJ1
 
 | | Puma BH3 foldamer in complex with Bcl-xL | | Descriptor: | ALPHA-BETA-PUMA BH3 FOLDAMER, BCL-2-LIKE PROTEIN 1 | | Authors: | Lee, E.F, Smith, B.J, Horne, W.S, Mayer, K.N, Evangelista, M, Colman, P.M, Gellman, S.H, Fairlie, W.D. | | Deposit date: | 2011-05-18 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Basis of Bcl-Xl Recognition by a Bh3-Mimetic Alpha-Beta-Peptide Generated Via Sequence-Based Design
Chembiochem, 12, 2011
|
|
5VAX
 
 | |
5VAU
 
 | |
2Y6W
 
 | | Structure of a Bcl-w dimer | | Descriptor: | BCL-2-LIKE PROTEIN 2, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL | | Authors: | Lee, E.F, Evangelista, M, Pettikiriarachchi, A, Dogovski, C, Perugini, M.A, Colman, P.M, Fairlie, W.D. | | Deposit date: | 2011-01-27 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a Bcl-W Domain-Swapped Dimer: Implications for the Function of Bcl-2 Family Proteins.
Structure, 19, 2011
|
|
3D7V
 
 | | Crystal structure of Mcl-1 in complex with an Mcl-1 selective BH3 ligand | | Descriptor: | Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Lee, E.F, Czabotar, P.E, Colman, P.M, Fairlie, W.D. | | Deposit date: | 2008-05-22 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A novel BH3 ligand that selectively targets Mcl-1 reveals that apoptosis can proceed without Mcl-1 degradation.
J.Cell Biol., 180, 2008
|
|
5VAY
 
 | |
6DCN
 
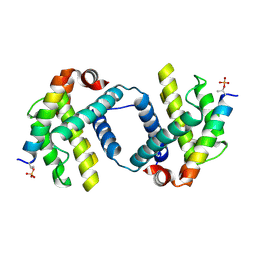 | | Bcl-xL complex with Beclin 1 BH3 domain T108pThr | | Descriptor: | BCL-xl protein, Beclin-1 | | Authors: | Lee, E.F, Smith, B.J, Smith, N.A, Yao, S, Fairlie, W.D. | | Deposit date: | 2018-05-07 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Structural insights into BCL2 pro-survival protein interactions with the key autophagy regulator BECN1 following phosphorylation by STK4/MST1.
Autophagy, 15, 2019
|
|
6DCO
 
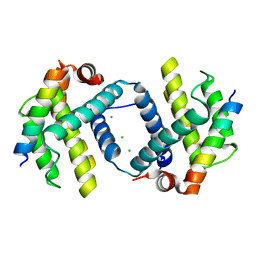 | | Bcl-xL complex with Beclin 1 BH3 domain T108D | | Descriptor: | Bcl-2-like protein 1,Bcl-2-like protein 1, Beclin-1, CHLORIDE ION | | Authors: | Lee, E.F, Smith, B.J, Smith, N.A, Yao, S, Fairlie, W.D. | | Deposit date: | 2018-05-07 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structural insights into BCL2 pro-survival protein interactions with the key autophagy regulator BECN1 following phosphorylation by STK4/MST1.
Autophagy, 15, 2019
|
|
3QBR
 
 | | BakBH3 in complex with sjA | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Bcl-2 homologous antagonist/killer, SJCHGC06286 protein | | Authors: | Lee, E.F, Clarke, O.B, Fairlie, W.D, Colman, P.M, Evangelista, M, Feng, Z, Speed, T.P, Tchoubrieva, E, Strasser, A, Kalinna, B. | | Deposit date: | 2011-01-13 | | Release date: | 2011-04-13 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Discovery and molecular characterization of a Bcl-2-regulated cell death pathway in schistosomes.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2PKO
 
 | |
7LYG
 
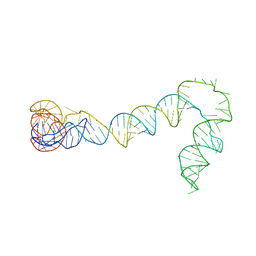 | |
7LYF
 
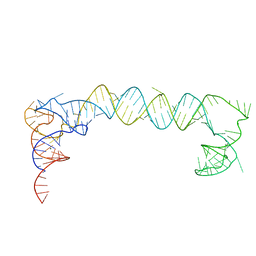 | |
4X2Y
 
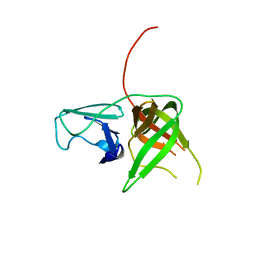 | | Crystal structure of a chimeric Murine Norovirus NS6 protease (inactive C139A mutant) in which the P4-P4 prime residues of the cleavage junction in the extended C-terminus have been replaced by the corresponding residues from the NS2-3 junction. | | Descriptor: | NS6 Protease,NS6 Protease | | Authors: | Leen, E.N, Pfeil, M.-P, Fernandes, H, Curry, S. | | Deposit date: | 2014-11-27 | | Release date: | 2015-02-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.417 Å) | | Cite: | Structure determination of Murine Norovirus NS6 proteases with C-terminal extensions designed to probe protease-substrate interactions.
Peerj, 3, 2015
|
|
4X2X
 
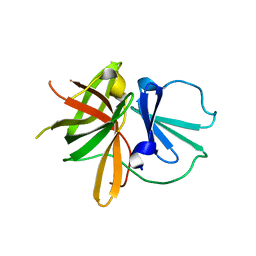 | |
2M4G
 
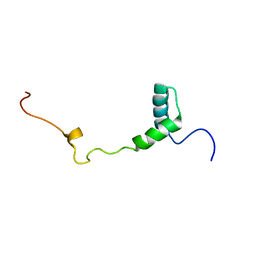 | | Solution structure of the Core Domain (11-85) of the Murine Norovirus VPg protein | | Descriptor: | Murine Norovirus VPg protein | | Authors: | Leen, E.N, Kwok, R, Birtley, J.R, Prater, S.N, Simpson, P.J, Matthews, S, Marchant, J, Curry, S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of the Compact Helical Core Domains of Feline Calicivirus and Murine Norovirus VPg Proteins.
J.Virol., 87, 2013
|
|
4ASH
 
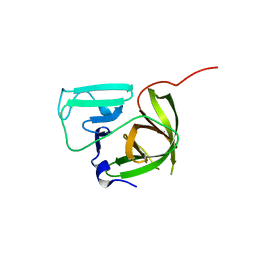 | |
3IO8
 
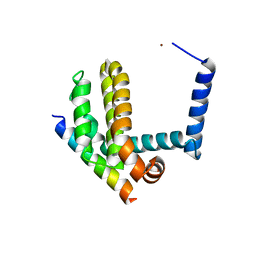 | | BimL12F in complex with Bcl-xL | | Descriptor: | Bcl-2-like protein 1, Bcl-2-like protein 11, ZINC ION | | Authors: | Colman, P.M, Lee, E.F, Fairlie, W.D, Smith, B.J, Czabotar, P.E, Yang, H, Sleebs, B.E, Lessene, G. | | Deposit date: | 2009-08-14 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational changes in Bcl-2 pro-survival proteins determine their capacity to bind ligands.
J.Biol.Chem., 284, 2009
|
|
3IO9
 
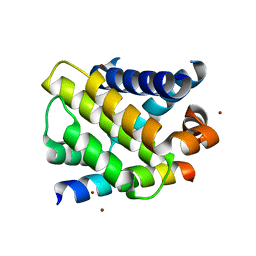 | | BimL12Y in complex with Mcl-1 | | Descriptor: | Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Czabotar, P.E, Lee, E.F, Yang, H, Sleebs, B.E, Lessene, G, Colman, P.M, Smith, B.J, Fairlie, W.D. | | Deposit date: | 2009-08-14 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational changes in Bcl-2 pro-survival proteins determine their capacity to bind ligands.
J.Biol.Chem., 284, 2009
|
|
4LWJ
 
 | |
4U66
 
 | |
