7U14
 
 | | TMEM106B(120-254) singlet amyloid fibril from frontotemporal lobar degeneration with TDP-43 pathology (FTLD-TDP) type C (case 8) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | 著者 | Fitzpatrick, A.W.P, Stowell, M.H.B, Chang, A, Xiang, X, Wang, J, Lee, C, Arakhamia, T, Simjanoska, M, Wang, C, Carlomagno, Y, Zhang, G, Dhingra, S, Thierry, M, Perneel, J, Heeman, B, Forgrave, L.M, DeTure, M, DeMarco, M.L, Cook, C.N, Rademakers, R, Dickson, D, Petrucelli, L, Mackenzie, I.R.A. | | 登録日 | 2022-02-19 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Homotypic fibrillization of TMEM106B across diverse neurodegenerative diseases.
Cell, 185, 2022
|
|
7EXP
 
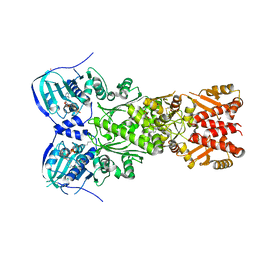 | | Crystal structure of zebrafish TRAP1 with AMPPNP and MitoQ | | 分子名称: | 2,3-dimethoxy-5-methyl-6-[10-(triphenyl-$l^{5}-phosphanyl)decyl]cyclohexa-2,5-diene-1,4-dione, COBALT (II) ION, MAGNESIUM ION, ... | | 著者 | Lee, H, Yoon, N.G, Kang, B.H, Lee, C. | | 登録日 | 2021-05-28 | | 公開日 | 2022-01-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.297 Å) | | 主引用文献 | Mitoquinone Inactivates Mitochondrial Chaperone TRAP1 by Blocking the Client Binding Site.
J.Am.Chem.Soc., 143, 2021
|
|
7MCI
 
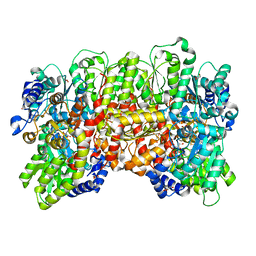 | | MoFe protein from Azotobacter vinelandii with a sulfur-replenished cofactor | | 分子名称: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | 著者 | Kang, W, Lee, C, Hu, Y, Ribbe, M.W. | | 登録日 | 2021-04-02 | | 公開日 | 2022-05-18 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Evidence of substrate binding and product release via belt-sulfur mobilization of the nitrogenase cofactor
Nat Catal, 5, 2022
|
|
2ZXX
 
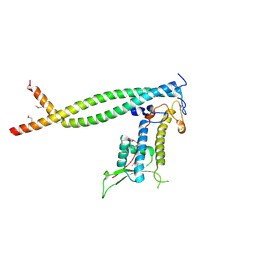 | | Crystal structure of Cdt1/geminin complex | | 分子名称: | DNA replication factor Cdt1, Geminin | | 著者 | Cho, Y, Lee, C, Hong, B.S, Choi, J.M. | | 登録日 | 2009-01-08 | | 公開日 | 2009-02-17 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis for inhibition of the replication licensing factor Cdt1 by geminin
Nature, 430, 2004
|
|
1CE3
 
 | |
2MBC
 
 | | Solution Structure of human holo-PRL-3 in complex with vanadate | | 分子名称: | Protein tyrosine phosphatase type IVA 3 | | 著者 | Jeong, K, Kang, D, Kim, J, Shin, S, Jin, B, Lee, C, Kim, E, Jeon, Y.H, Kim, Y. | | 登録日 | 2013-07-29 | | 公開日 | 2013-10-09 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and backbone dynamics of vanadate-bound PRL-3: comparison of 15N nuclear magnetic resonance relaxation profiles of free and vanadate-bound PRL-3.
Biochemistry, 53, 2014
|
|
5E1J
 
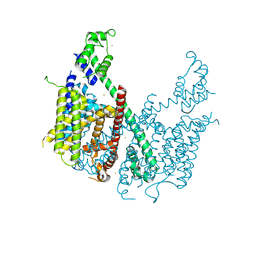 | | Structure of voltage-gated two-pore channel TPC1 from Arabidopsis thaliana | | 分子名称: | BARIUM ION, CALCIUM ION, Two pore calcium channel protein 1 | | 著者 | Guo, J, Zeng, W, Chen, Q, Lee, C, Chen, L, Yang, Y, Jiang, Y. | | 登録日 | 2015-09-29 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.308 Å) | | 主引用文献 | Structure of the voltage-gated two-pore channel TPC1 from Arabidopsis thaliana.
Nature, 531, 2016
|
|
1G4B
 
 | | CRYSTAL STRUCTURES OF THE HSLVU PEPTIDASE-ATPASE COMPLEX REVEAL AN ATP-DEPENDENT PROTEOLYSIS MECHANISM | | 分子名称: | ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | 著者 | Wang, J, Song, J.J, Franklin, M.C, Kamtekar, S, Im, Y.J, Rho, S.H, Seong, I.S, Lee, C.S, Chung, C.H, Eom, S.H. | | 登録日 | 2000-10-26 | | 公開日 | 2001-02-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (7 Å) | | 主引用文献 | Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism.
Structure, 9, 2001
|
|
1G4A
 
 | | CRYSTAL STRUCTURES OF THE HSLVU PEPTIDASE-ATPASE COMPLEX REVEAL AN ATP-DEPENDENT PROTEOLYSIS MECHANISM | | 分子名称: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, ATP-DEPENDENT PROTEASE HSLV | | 著者 | Wang, J, Song, J.J, Franklin, M.C, Kamtekar, S, Im, Y.J, Rho, S.H, Seong, I.S, Lee, C.S, Chung, C.H, Eom, S.H. | | 登録日 | 2000-10-26 | | 公開日 | 2001-02-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism.
Structure, 9, 2001
|
|
2L01
 
 | | Solution NMR Structure of protein BVU3908 from Bacteroides vulgatus, Northeast Structural Genomics Consortium Target BvR153 | | 分子名称: | Uncharacterized protein | | 著者 | Eletsky, A, Lee, C, Wang, K, Ciccosanti, T.B, Hamilton, R, Acton, J.B, Xiao, G.B, Everett, J.K, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2010-06-29 | | 公開日 | 2010-08-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution NMR Structure of protein BVU3908 from Bacteroides vulgatus
To be Published
|
|
1XZY
 
 | | Solution structure of the P30-trans form of Alpha Hemoglobin Stabilizing Protein (AHSP) | | 分子名称: | Alpha-hemoglobin stabilizing protein | | 著者 | Gell, D.A, Feng, L, Zhou, S, Kong, Y, Lee, C, Weiss, M.J, Shi, Y, Mackay, J.P. | | 登録日 | 2004-11-12 | | 公開日 | 2004-12-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular mechanism of AHSP-mediated stabilization of alpha-hemoglobin
Cell(Cambridge,Mass.), 119, 2004
|
|
7C7B
 
 | | Crystal structure of human TRAP1 with SJT009 | | 分子名称: | 2-azanyl-9-[(6-bromanyl-1,3-benzodioxol-5-yl)methyl]-6-chloranyl-purin-8-ol, Heat shock protein 75 kDa, mitochondrial | | 著者 | Kim, D, Yang, S, Yoon, N.G, Park, E, Kim, S.Y, Kang, B.H, Lee, C, Kang, S. | | 登録日 | 2020-05-24 | | 公開日 | 2021-05-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Design and Synthesis of TRAP1 Selective Inhibitors: H-Bonding with Asn171 Residue in TRAP1 Increases Paralog Selectivity.
Acs Med.Chem.Lett., 12, 2021
|
|
7C7C
 
 | | Crystal structure of human TRAP1 with SJT104 | | 分子名称: | 2-azanyl-9-[(4-bromanyl-2-fluoranyl-phenyl)methyl]-6-chloranyl-purin-8-ol, Heat shock protein 75 kDa, mitochondrial | | 著者 | Kim, D, Yang, S, Yoon, N.G, Park, E, Kim, S.Y, Kang, B.H, Lee, C, Kang, S. | | 登録日 | 2020-05-24 | | 公開日 | 2021-05-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Design and Synthesis of TRAP1 Selective Inhibitors: H-Bonding with Asn171 Residue in TRAP1 Increases Paralog Selectivity.
Acs Med.Chem.Lett., 12, 2021
|
|
5YK6
 
 | | Crystal Structure of Mmm1 | | 分子名称: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, Maintenance of mitochondrial morphology protein 1 | | 著者 | Jeong, H, Park, J, Lee, C. | | 登録日 | 2017-10-12 | | 公開日 | 2018-01-10 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structures of Mmm1 and Mdm12-Mmm1 reveal mechanistic insight into phospholipid trafficking at ER-mitochondria contact sites.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5YK7
 
 | | Crystal Structure of Mdm12-Mmm1 complex | | 分子名称: | Maintenance of mitochondrial morphology protein 1, Mitochondrial distribution and morphology protein 12, PHOSPHATE ION | | 著者 | Jeong, H, Park, J, Lee, C. | | 登録日 | 2017-10-12 | | 公開日 | 2018-01-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.799 Å) | | 主引用文献 | Crystal structures of Mmm1 and Mdm12-Mmm1 reveal mechanistic insight into phospholipid trafficking at ER-mitochondria contact sites.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4MLG
 
 | | Structure of RS223-Beta-xylosidase | | 分子名称: | Beta-xylosidase, CALCIUM ION, SULFATE ION | | 著者 | Jordan, D, Braker, J, Wagschal, K, Lee, C, Dubrovska, I, Anderson, S, Wawrzak, Z. | | 登録日 | 2013-09-06 | | 公開日 | 2014-09-17 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of RS223-Beta-xylosidase
To be Published
|
|
2J3K
 
 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Ternary Complex II | | 分子名称: | (2E,4R)-4-HYDROXYNON-2-ENAL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-dependent oxidoreductase 2-alkenal reductase | | 著者 | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | 登録日 | 2006-08-22 | | 公開日 | 2006-10-05 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Mechanistic and structural studies of apoform, binary, and ternary complexes of the Arabidopsis alkenal double bond reductase At5g16970.
J. Biol. Chem., 281, 2006
|
|
2J3I
 
 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Binary Complex | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-DEPENDENT OXIDOREDUCTASE P1 | | 著者 | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | 登録日 | 2006-08-21 | | 公開日 | 2006-10-05 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Mechanistic and Structural Studies of Apoform, Binary, and Ternary Complexes of the Arabidopsis Alkenal Double Bond Reductase at5G16970.
J.Biol.Chem., 281, 2006
|
|
2J3H
 
 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Apo form | | 分子名称: | NADP-DEPENDENT OXIDOREDUCTASE P1 | | 著者 | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | 登録日 | 2006-08-21 | | 公開日 | 2006-10-05 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Mechanistic and Structural Studies of Apoform, Binary, and Ternary Complexes of the Arabidopsis Alkenal Double Bond Reductase at5G16970.
J.Biol.Chem., 281, 2006
|
|
2J3J
 
 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Ternary Complex I | | 分子名称: | 4'-HYDROXYCINNAMIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-dependent oxidoreductase 2-alkenal reductase | | 著者 | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | 登録日 | 2006-08-21 | | 公開日 | 2006-10-05 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Mechanistic and structural studies of apoform, binary, and ternary complexes of the Arabidopsis alkenal double bond reductase At5g16970.
J. Biol. Chem., 281, 2006
|
|
5GYD
 
 | | Crystal Structure of Mdm12 | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Mitochondrial distribution and morphology protein 12 | | 著者 | Jeong, H, Park, J, Lee, C. | | 登録日 | 2016-09-22 | | 公開日 | 2016-11-16 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.106 Å) | | 主引用文献 | Crystal structure of Mdm12 reveals the architecture and dynamic organization of the ERMES complex
EMBO Rep., 17, 2016
|
|
5GYK
 
 | | Crystal Structure of Mdm12-deletion mutant | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Mitochondrial distribution and morphology protein 12 | | 著者 | Jeong, H, Park, J, Lee, C. | | 登録日 | 2016-09-22 | | 公開日 | 2016-11-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.596 Å) | | 主引用文献 | Crystal structure of Mdm12 reveals the architecture and dynamic organization of the ERMES complex
EMBO Rep., 17, 2016
|
|
2MXD
 
 | | Solution structure of VPg of porcine sapovirus | | 分子名称: | Viral protein genome-linked | | 著者 | Kim, J, Hwang, H, Min, H, Yun, H, Cho, K, Pelton, J.G, Wemmer, D.E, Lee, C. | | 登録日 | 2014-12-24 | | 公開日 | 2015-04-15 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the porcine sapovirus VPg core reveals a stable three-helical bundle with a conserved surface patch.
Biochem.Biophys.Res.Commun., 459, 2015
|
|
