7DVN
 
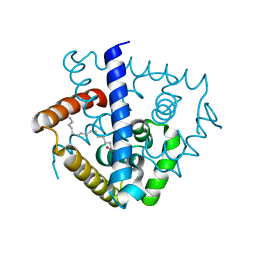 | | Crystal structure of a MarR family protein in complex with a lipid-like effector molecule from the psychrophilic bacterium Paenisporosarcina sp. TG-14 | | Descriptor: | MarR family transcriptional regulator, PALMITIC ACID | | Authors: | Lee, C.W, Hwang, J, Do, H, Lee, J.H. | | Deposit date: | 2021-01-14 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a MarR family protein from the psychrophilic bacterium Paenisporosarcina sp. TG-14 in complex with a lipid-like molecule.
Iucrj, 8, 2021
|
|
3Q6N
 
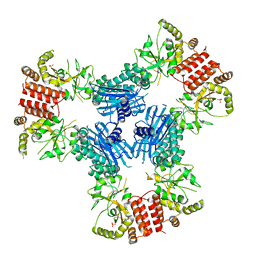 | | Crystal Structure of Human MC-HSP90 in P21 space group | | Descriptor: | Heat shock protein HSP 90-alpha, SULFATE ION | | Authors: | Lee, C.C, Lin, T.W, Ko, T.P, Wang, A.H.-J. | | Deposit date: | 2011-01-03 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | The hexameric structures of human heat shock protein 90
Plos One, 6, 2011
|
|
3Q6M
 
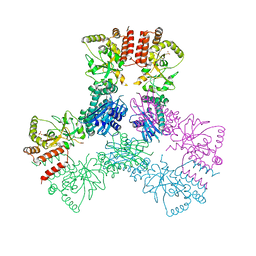 | | Crystal Structure of Human MC-HSP90 in C2221 Space Group | | Descriptor: | Heat shock protein HSP 90-alpha, SULFATE ION | | Authors: | Lee, C.C, Lin, T.W, Ko, T.P, Wang, A.H.-J. | | Deposit date: | 2011-01-03 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The hexameric structures of human heat shock protein 90
Plos One, 6, 2011
|
|
4PX6
 
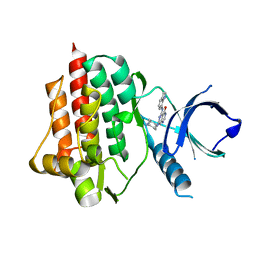 | | SYK catalytic domain in complex with a potent pyridopyrimidinone inhibitor | | Descriptor: | 7-{[(1R,2S)-2-aminocyclohexyl]amino}-5-(1H-indol-7-ylamino)pyrido[4,3-d]pyrimidin-4(3H)-one, Tyrosine-protein kinase SYK | | Authors: | Lee, C.C. | | Deposit date: | 2014-03-21 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Syk inhibitors with high potency in presence of blood.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
5U6O
 
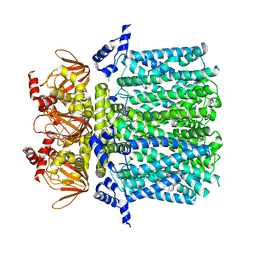 | |
5HXT
 
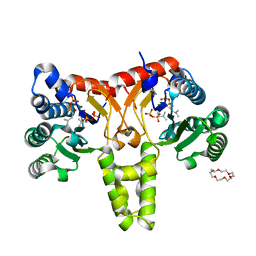 | | Crystal Structure of Z,Z-Farnesyl Diphosphate Synthase (D71M, E75A and H103Y Mutants) Complexed with IPP and DMSPP | | Descriptor: | (2Z,6Z)-farnesyl diphosphate synthase, chloroplastic, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, ... | | Authors: | Lee, C.C, Chan, Y.T, Wang, A.H.J. | | Deposit date: | 2016-01-31 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure and Potential Head-to-Middle Condensation Function of a Z,Z-Farnesyl Diphosphate Synthase
Acs Omega, 2, 2017
|
|
6ABT
 
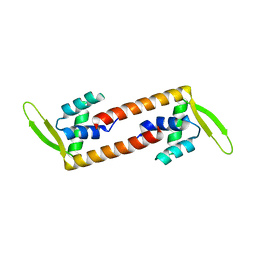 | | Crystal structure of transcription factor from Listeria monocytogenes | | Descriptor: | PadR family transcriptional regulator | | Authors: | Lee, C, Hong, M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based molecular characterization and regulatory mechanism of the LftR transcription factor from Listeria monocytogenes: Conformational flexibilities and a ligand-induced regulatory mechanism.
Plos One, 14, 2019
|
|
6JP7
 
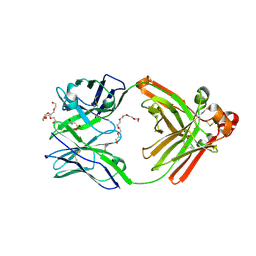 | | Human antibody 32D6 Fab in complex with PEG | | Descriptor: | (2R)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, HEXAETHYLENE GLYCOL, immunoglobulin Fab heavy chain, ... | | Authors: | Lee, C.C, Ko, T.P, Lin, L.L, Wang, A.H.J. | | Deposit date: | 2019-03-26 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.909 Å) | | Cite: | Structural basis of polyethylene glycol recognition by antibody.
J.Biomed.Sci., 27, 2020
|
|
6JU0
 
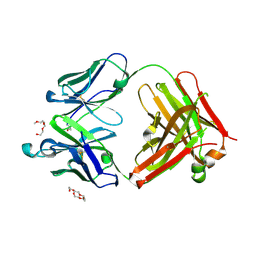 | | Mouse antibody 3.3 Fab in complex with PEG | | Descriptor: | 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Fab heavy chain, ... | | Authors: | Lee, C.C, Ko, T.P, Lin, L.L, Wang, A.H.J. | | Deposit date: | 2019-04-12 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural basis of polyethylene glycol recognition by antibody.
J.Biomed.Sci., 27, 2020
|
|
6JWC
 
 | | Mouse antibody 2B5 Fab in complex with PEG | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, DI(HYDROXYETHYL)ETHER, DODECAETHYLENE GLYCOL, ... | | Authors: | Lee, C.C, Ko, T.P, Su, Y.C, Lin, L.L, Roffler, S.R, Wang, A.H.J. | | Deposit date: | 2019-04-19 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Structural basis of polyethylene glycol recognition by antibody.
J.Biomed.Sci., 27, 2020
|
|
5HXN
 
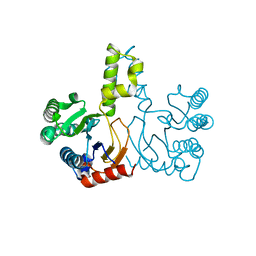 | | Crystal Structure of Z,Z-Farnesyl Diphosphate Synthase (D71M and E75A mutants) from the Wild Tomato Solanum habrochaites | | Descriptor: | (2Z,6Z)-farnesyl diphosphate synthase, chloroplastic | | Authors: | Lee, C.C, Chan, Y.T, Wang, A.H.J. | | Deposit date: | 2016-01-31 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure and Potential Head-to-Middle Condensation Function of aZ,Z-Farnesyl Diphosphate Synthase.
Acs Omega, 2, 2017
|
|
5HXO
 
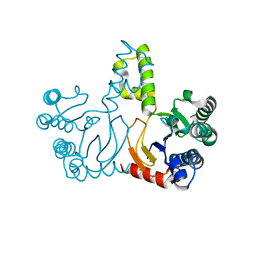 | | Crystal Structure of Z,Z-Farnesyl Diphosphate Synthase with D71M, E75A and H103Y Mutants | | Descriptor: | (2Z,6Z)-farnesyl diphosphate synthase, chloroplastic | | Authors: | Lee, C.C, Chan, Y.T, Wang, A.H.J. | | Deposit date: | 2016-01-31 | | Release date: | 2017-04-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure and Potential Head-to-Middle Condensation Function of aZ,Z-Farnesyl Diphosphate Synthase.
Acs Omega, 2, 2017
|
|
2L14
 
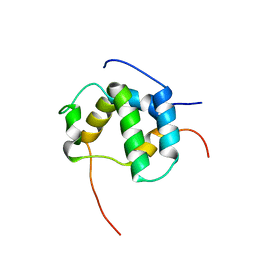 | | Structure of CBP nuclear coactivator binding domain in complex with p53 TAD | | Descriptor: | CREB-binding protein, Cellular tumor antigen p53 | | Authors: | Lee, C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2010-07-22 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the p53 transactivation domain in complex with the nuclear receptor coactivator binding domain of CREB binding protein.
Biochemistry, 49, 2010
|
|
1T1T
 
 | | Solution Structure of Kurtoxin | | Descriptor: | Kurtoxin | | Authors: | Lee, C.W, Min, H.J, Cho, E.M, Kohno, T, Eu, Y.J, Kim, J.I. | | Deposit date: | 2004-04-18 | | Release date: | 2005-06-28 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of kurtoxin
To be Published
|
|
3NOW
 
 | | UNC-45 from Drosophila melanogaster | | Descriptor: | UNC-45 protein, SD10334p | | Authors: | Lee, C.F, Hauenstein, A.V, Fleming, J.K, Gasper, W.C, Engelke, V, Banumathi, S, Bernstein, S.I, Huxford, T. | | Deposit date: | 2010-06-25 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.992 Å) | | Cite: | X-ray Crystal Structure of the UCS Domain-Containing UNC-45 Myosin Chaperone from Drosophila melanogaster.
Structure, 19, 2011
|
|
2IJM
 
 | |
1LA4
 
 | | Solution Structure of SGTx1 | | Descriptor: | SGTx1 | | Authors: | Lee, C.W, Roh, S.H, Kim, S, Endoh, H, Kodera, Y, Maeda, T, Swartz, K.J, Kim, J.I. | | Deposit date: | 2002-03-28 | | Release date: | 2003-11-11 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Functional Characterization of SGTx1, a Modifier of Kv2.1 Channel Gating
Biochemistry, 43, 2004
|
|
2LKP
 
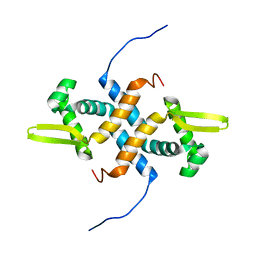 | | solution structure of apo-NmtR | | Descriptor: | HTH-type transcriptional regulator NmtR | | Authors: | Lee, C, Giedroc, D. | | Deposit date: | 2011-10-18 | | Release date: | 2012-04-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Mycobacterium tuberculosis NmtR in the Apo State: Insights into Ni(II)-Mediated Allostery.
Biochemistry, 51, 2012
|
|
3TNU
 
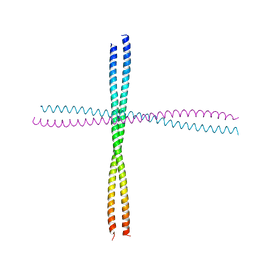 | | Heterocomplex of coil 2B domains of human intermediate filament proteins, keratin 5 (KRT5) and keratin 14 (KRT14) | | Descriptor: | Keratin, type I cytoskeletal 14, type II cytoskeletal 5 | | Authors: | Lee, C.H, Kim, M.S, Leahy, D.J, Coulombe, P.A. | | Deposit date: | 2011-09-02 | | Release date: | 2012-06-20 | | Last modified: | 2012-08-01 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Structural basis for heteromeric assembly and perinuclear organization of keratin filaments.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5H20
 
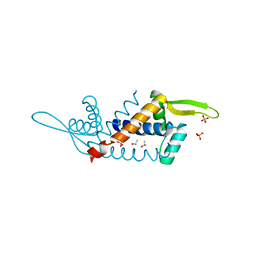 | | X-ray structure of PadR-like Transcription factor from bacteroid fragilis | | Descriptor: | ISOPROPYL ALCOHOL, PHOSPHATE ION, Putative PadR-family transcriptional regulatory protein, ... | | Authors: | Lee, C, Hong, M. | | Deposit date: | 2016-10-13 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional analysis of BF2549, a PadR-like transcription factor from Bacteroides fragilis.
Biochem. Biophys. Res. Commun., 483, 2017
|
|
5XEF
 
 | | Crystal structure of flagellar chaperone from bacteria | | Descriptor: | Flagellar protein fliS | | Authors: | Lee, C, Hong, M. | | Deposit date: | 2017-04-05 | | Release date: | 2018-06-27 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the flagellar chaperone FliS from Bacillus cereus and an invariant proline critical for FliS dimerization and flagellin recognition
Biochem. Biophys. Res. Commun., 487, 2017
|
|
5GWG
 
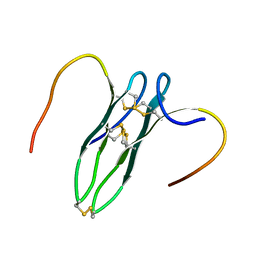 | | Solution structure of rattusin | | Descriptor: | Defensin alpha-related sequence 1 | | Authors: | Lee, C.W, Min, H.J. | | Deposit date: | 2016-09-11 | | Release date: | 2017-04-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Rattusin structure reveals a novel defensin scaffold formed by intermolecular disulfide exchanges
Sci Rep, 7, 2017
|
|
2L6I
 
 | |
2HIW
 
 | |
7C8T
 
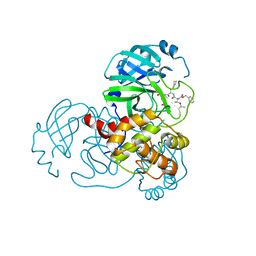 | | Complex Structure of SARS-CoV-2 3CL Protease with TG-0205221 | | Descriptor: | 3C-like proteinase, N-[(BENZYLOXY)CARBONYL]-O-(TERT-BUTYL)-L-THREONYL-3-CYCLOHEXYL-N-[(1S)-2-HYDROXY-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}ETHYL]-L-ALANINAMIDE | | Authors: | Lee, C.C, Wang, A.H.J, Kuo, C.J, Liang, P.H. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Complex Structures and Cellular Activities of the Potent SARS-CoV-2 3CLpro Inhibitors Guiding Drug Discovery Against COVID-19
To Be Published
|
|
