7UK5
 
 | |
7UKD
 
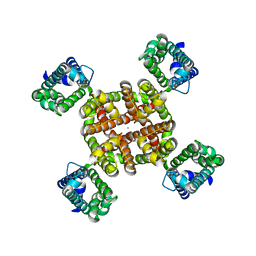 | | Human Kv4.2-KChIP2 channel complex in an inactivated state, class 2, transmembrane region | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Activation and closed-state inactivation mechanisms of the human voltage-gated K V 4 channel complexes.
Mol.Cell, 82, 2022
|
|
7UKF
 
 | | Human Kv4.2-KChIP2 channel complex in a putative resting state, transmembrane region | | Descriptor: | MERCURY (II) ION, POTASSIUM ION, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Activation and closed-state inactivation mechanisms of the human voltage-gated K V 4 channel complexes.
Mol.Cell, 82, 2022
|
|
7UKC
 
 | | Human Kv4.2-KChIP2 channel complex in an inactivated state, class 1, transmembrane region | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Zhao, H, Dai, Y, Lee, C.H. | | Deposit date: | 2022-04-01 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Activation and closed-state inactivation mechanisms of the human voltage-gated K V 4 channel complexes.
Mol.Cell, 82, 2022
|
|
5U86
 
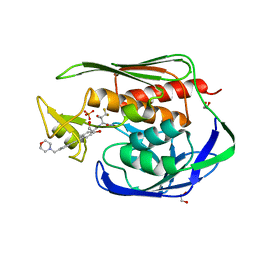 | | Structure of the Aquifex aeolicus LpxC/LPC-069 complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, N-[(2S,3S)-4,4-difluoro-3-hydroxy-1-(hydroxyamino)-3-methyl-1-oxobutan-2-yl]-4-({4-[(morpholin-4-yl)methyl]phenyl}ethynyl)benzamide, ... | | Authors: | Najeeb, J, Lee, C.-J, Zhou, P. | | Deposit date: | 2016-12-13 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Curative Treatment of Severe Gram-Negative Bacterial Infections by a New Class of Antibiotics Targeting LpxC.
MBio, 8, 2017
|
|
5XQH
 
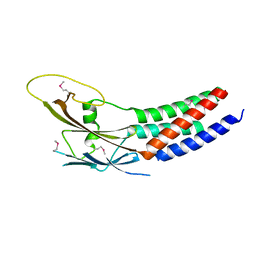 | | Crystal structure of truncated human Rogdi | | Descriptor: | Protein rogdi homolog | | Authors: | Lee, H, Lee, C. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The crystal structure of human Rogdi provides insight into the causes of Kohlschutter-Tonz Syndrome
Sci Rep, 7, 2017
|
|
6K1D
 
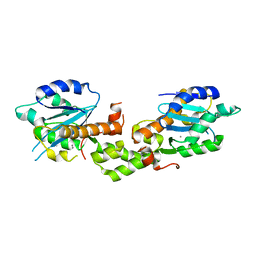 | | Crystal structure of EXD2 exonuclease domain soaked in Mn and GMP | | Descriptor: | Exonuclease 3'-5' domain-containing protein 2, MANGANESE (II) ION | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of human EXD2 reveals a chimeric 3' to 5' exonuclease domain that discriminates substrates via metal coordination.
Nucleic Acids Res., 47, 2019
|
|
6K1C
 
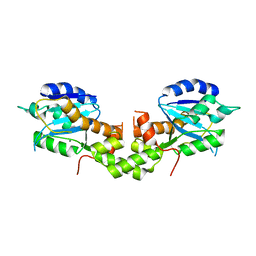 | | Crystal structure of EXD2 exonuclease domain soaked in Mg and dGMP | | Descriptor: | Exonuclease 3'-5' domain-containing protein 2, MAGNESIUM ION | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The structure of human EXD2 reveals a chimeric 3' to 5' exonuclease domain that discriminates substrates via metal coordination.
Nucleic Acids Res., 47, 2019
|
|
6K18
 
 | | Crystal structure of EXD2 exonuclease domain soaked in Mn | | Descriptor: | Exonuclease 3'-5' domain-containing protein 2, MANGANESE (II) ION | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | The structure of human EXD2 reveals a chimeric 3' to 5' exonuclease domain that discriminates substrates via metal coordination.
Nucleic Acids Res., 47, 2019
|
|
6K1A
 
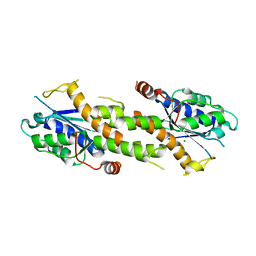 | | Crystal structure of EXD2 exonuclease domain soaked in Mn and Mg | | Descriptor: | Exonuclease 3'-5' domain-containing protein 2, MAGNESIUM ION, MANGANESE (II) ION | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | The structure of human EXD2 reveals a chimeric 3' to 5' exonuclease domain that discriminates substrates via metal coordination.
Nucleic Acids Res., 47, 2019
|
|
6K17
 
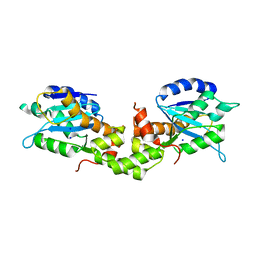 | | Crystal structure of EXD2 exonuclease domain | | Descriptor: | Exonuclease 3'-5' domain-containing protein 2, SODIUM ION | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | The structure of human EXD2 reveals a chimeric 3' to 5' exonuclease domain that discriminates substrates via metal coordination.
Nucleic Acids Res., 47, 2019
|
|
6K1E
 
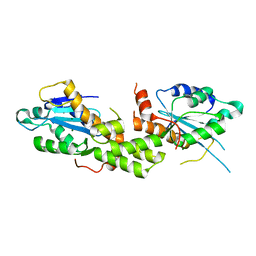 | | Crystal structure of EXD2 exonuclease domain soaked in Mg and GMP | | Descriptor: | Exonuclease 3'-5' domain-containing protein 2, MAGNESIUM ION | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of human EXD2 reveals a chimeric 3' to 5' exonuclease domain that discriminates substrates via metal coordination.
Nucleic Acids Res., 47, 2019
|
|
6K1B
 
 | | Crystal structure of EXD2 exonuclease domain soaked in Mn and dGMP | | Descriptor: | Exonuclease 3'-5' domain-containing protein 2, MANGANESE (II) ION | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | The structure of human EXD2 reveals a chimeric 3' to 5' exonuclease domain that discriminates substrates via metal coordination.
Nucleic Acids Res., 47, 2019
|
|
1XZY
 
 | | Solution structure of the P30-trans form of Alpha Hemoglobin Stabilizing Protein (AHSP) | | Descriptor: | Alpha-hemoglobin stabilizing protein | | Authors: | Gell, D.A, Feng, L, Zhou, S, Kong, Y, Lee, C, Weiss, M.J, Shi, Y, Mackay, J.P. | | Deposit date: | 2004-11-12 | | Release date: | 2004-12-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Molecular mechanism of AHSP-mediated stabilization of alpha-hemoglobin
Cell(Cambridge,Mass.), 119, 2004
|
|
6KBN
 
 | | Crystal structure of Vac8 (del 19-33) bound to Atg13 | | Descriptor: | Autophagy-related protein 13, Vacuolar protein 8 | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-06-25 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Quaternary structures of Vac8 differentially regulate the Cvt and PMN pathways.
Autophagy, 16, 2020
|
|
8D36
 
 | |
7F94
 
 | | Structure of C-terminal truncated connexin43/Cx43/GJA1 gap junction intercellular channel with two conformationally different hemichannels | | Descriptor: | A C-terminal deletion mutant of gap junction alpha-1 protein (Cx43-M257) | | Authors: | Lee, H.J, Cha, H.J, Jeong, H, Lee, S.N, Lee, C.W, Woo, J.S. | | Deposit date: | 2021-07-03 | | Release date: | 2022-07-06 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Conformational changes in the human Cx43/GJA1 gap junction channel visualized using cryo-EM.
Nat Commun, 14, 2023
|
|
4AU5
 
 | | Structure of the NhaA dimer, crystallised at low pH | | Descriptor: | DODECYL-ALPHA-D-MALTOSIDE, NA(+)/H(+) ANTIPORTER NHAA, SULFATE ION | | Authors: | Drew, D, Lee, C, Iwata, S, Cameron, A.D. | | Deposit date: | 2012-05-14 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.696 Å) | | Cite: | Crystal structure of the sodium-proton antiporter NhaA dimer and new mechanistic insights.
J. Gen. Physiol., 144, 2014
|
|
6K19
 
 | | Crystal structure of EXD2 exonuclease domain soaked in Mg | | Descriptor: | Exonuclease 3'-5' domain-containing protein 2, MAGNESIUM ION | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-05-10 | | Release date: | 2019-05-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | The structure of human EXD2 reveals a chimeric 3' to 5' exonuclease domain that discriminates substrates via metal coordination.
Nucleic Acids Res., 47, 2019
|
|
5DRP
 
 | | Structure of the AaLpxC/LPC-023 Complex | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, N~2~-{4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]benzoyl}-N-hydroxy-L-isoleucinamide, ... | | Authors: | Najeeb, J, Lee, C.-J, Zhou, P. | | Deposit date: | 2015-09-16 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Drug design from the cryptic inhibitor envelope.
Nat Commun, 7, 2016
|
|
2VGP
 
 | | Crystal structure of Aurora B kinase in complex with a aminothiazole inhibitor | | Descriptor: | 4-[(5-bromo-1,3-thiazol-2-yl)amino]-N-methylbenzamide, INNER CENTROMERE PROTEIN A, SERINE/THREONINE-PROTEIN KINASE 12-A | | Authors: | Andersen, C.B, Wan, Y, Chang, J.W, Lee, C, Liu, Y, Sessa, F, Villa, F, Nallan, L, Musacchio, A, Gray, N.S. | | Deposit date: | 2007-11-15 | | Release date: | 2008-02-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Selective Aminothiazole Aurora Kinase Inhibitors
Acs Chem.Biol., 3, 2008
|
|
3DZD
 
 | | Crystal structure of sigma54 activator NTRC4 in the inactive state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SODIUM ION, Transcriptional regulator (NtrC family) | | Authors: | Batchelor, J.D, Doucleff, M, Lee, C.-J, Matsubara, K, De Carlo, S, Heideker, J, Lamers, M.M, Pelton, J.G, Wemmer, D.E. | | Deposit date: | 2008-07-29 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and regulatory mechanism of Aquifex aeolicus NtrC4: variability and evolution in bacterial transcriptional regulation.
J.Mol.Biol., 384, 2008
|
|
8DAO
 
 | | Crystal structure of SARS-CoV-2 spike stem fusion peptide in complex with neutralizing antibody COV44-79 | | Descriptor: | COV44-79 heavy chain constant domain, COV44-79 heavy chain variable domain, COV44-79 light chain constant domain, ... | | Authors: | Lin, T.H, Lee, C.C.D, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-06-13 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Broadly neutralizing antibodies target the coronavirus fusion peptide.
Science, 377, 2022
|
|
5XQI
 
 | | Crystal structure of full-length human Rogdi | | Descriptor: | Protein rogdi homolog | | Authors: | Lee, H, Lee, C. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of human Rogdi provides insight into the causes of Kohlschutter-Tonz Syndrome
Sci Rep, 7, 2017
|
|
7C7B
 
 | | Crystal structure of human TRAP1 with SJT009 | | Descriptor: | 2-azanyl-9-[(6-bromanyl-1,3-benzodioxol-5-yl)methyl]-6-chloranyl-purin-8-ol, Heat shock protein 75 kDa, mitochondrial | | Authors: | Kim, D, Yang, S, Yoon, N.G, Park, E, Kim, S.Y, Kang, B.H, Lee, C, Kang, S. | | Deposit date: | 2020-05-24 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design and Synthesis of TRAP1 Selective Inhibitors: H-Bonding with Asn171 Residue in TRAP1 Increases Paralog Selectivity.
Acs Med.Chem.Lett., 12, 2021
|
|
