4RSS
 
 | | Crystal structure of tyrosine-protein kinase SYK with an inhibitor | | Descriptor: | 1-[(3-methyl-1-{2-[(1,2,3-trimethyl-1H-indol-5-yl)amino]pyrimidin-4-yl}-1H-pyrazol-4-yl)methyl]azetidin-3-ol, Tyrosine-protein kinase SYK | | Authors: | Lee, B.I, Lee, S.J, Choi, J.-S. | | Deposit date: | 2014-11-11 | | Release date: | 2015-10-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Highly potent and selective pyrazolylpyrimidines as Syk kinase inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
1HYE
 
 | | CRYSTAL STRUCTURE OF THE MJ0490 GENE PRODUCT, THE FAMILY OF LACTATE/MALATE DEHYDROGENASE, DIMERIC STRUCTURE | | Descriptor: | L-LACTATE/MALATE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lee, B.I, Chang, C, Cho, S.-J, Suh, S.W. | | Deposit date: | 2001-01-19 | | Release date: | 2001-04-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the MJ0490 gene product of the hyperthermophilic archaebacterium Methanococcus jannaschii, a novel member of the lactate/malate family of dehydrogenases.
J.Mol.Biol., 307, 2001
|
|
1HYG
 
 | | Crystal structure of MJ0490 gene product, the family of lactate/malate dehydrogenase | | Descriptor: | L-LACTATE/MALATE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lee, B.I, Chang, C, Cho, S.-J, Suh, S.W. | | Deposit date: | 2001-01-19 | | Release date: | 2001-04-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the MJ0490 gene product of the hyperthermophilic archaebacterium Methanococcus jannaschii, a novel member of the lactate/malate family of dehydrogenases
J.Mol.Biol., 307, 2001
|
|
9JEJ
 
 | |
9IPW
 
 | |
5GNH
 
 | | Myotubularin-related protein 2 | | Descriptor: | Myotubularin-related protein 2, PHOSPHATE ION | | Authors: | Lee, B.I, Bong, S.M. | | Deposit date: | 2016-07-21 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of MTMR2
To Be Published
|
|
7CC7
 
 | |
3V6A
 
 | |
1J2Z
 
 | | Crystal structure of UDP-N-acetylglucosamine acyltransferase | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, L(+)-TARTARIC ACID, SULFATE ION, ... | | Authors: | Lee, B.I, Suh, S.W. | | Deposit date: | 2003-01-15 | | Release date: | 2004-01-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of UDP-N-acetylglucosamine acyltransferase from Helicobacter pylori
Proteins, 53, 2003
|
|
1J2Y
 
 | | Crystal structure of the type II 3-dehydroquinase | | Descriptor: | 1,3,4-TRIHYDROXY-5-OXO-CYCLOHEXANECARBOXYLIC ACID, 3-dehydroquinate dehydratase | | Authors: | Lee, B.I, Kwak, J.E, Suh, S.W. | | Deposit date: | 2003-01-15 | | Release date: | 2003-06-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the type II 3-dehydroquinase from Helicobacter pylori
Proteins, 51, 2003
|
|
6KEI
 
 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with 16-methoxy-11-methyl-6-[(pyridin-2-yl)methoxy]-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(16),3,5,7,9,13(17),14-heptaen-12-one | | Descriptor: | 16-methoxy-11-methyl-6-[(pyridin-2-yl)methoxy]-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(16),3,5,7,9,13(17),14-heptaen-12-one, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Lee, B.I, Park, T.H. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Synthesis and Structure-Activity Relationships of Aristoyagonine Derivatives as Brd4 Bromodomain Inhibitors with X-ray Co-Crystal Research.
Molecules, 26, 2021
|
|
1VDD
 
 | |
6KEC
 
 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with 4-ethoxy-5,16-dimethoxy-11-methyl-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(17),3,5,7,9,13,15-heptaen-12-one | | Descriptor: | 4-ethoxy-5,16-dimethoxy-11-methyl-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(17),3,5,7,9,13,15-heptaen-12-one, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Lee, B.I, Park, T.H. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Synthesis and Structure-Activity Relationships of Aristoyagonine Derivatives as Brd4 Bromodomain Inhibitors with X-ray Co-Crystal Research.
Molecules, 26, 2021
|
|
6KEH
 
 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with 6,16-dimethoxy-11-methyl-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(17),3,5,7,9,13,15-heptaen-12-one | | Descriptor: | 6,16-dimethoxy-11-methyl-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(17),3,5,7,9,13,15-heptaen-12-one, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Lee, B.I, Park, T.H. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | Synthesis and Structure-Activity Relationships of Aristoyagonine Derivatives as Brd4 Bromodomain Inhibitors with X-ray Co-Crystal Research.
Molecules, 26, 2021
|
|
6KEJ
 
 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with 6-[2-(diethylamino)ethoxy]-16-methoxy-11-methyl-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(17),3,5,7,9,13,15-heptaen-12-one | | Descriptor: | 6-[2-(diethylamino)ethoxy]-16-methoxy-11-methyl-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(17),3,5,7,9,13,15-heptaen-12-one, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Lee, B.I, Park, T.H. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis and Structure-Activity Relationships of Aristoyagonine Derivatives as Brd4 Bromodomain Inhibitors with X-ray Co-Crystal Research.
Molecules, 26, 2021
|
|
6KEK
 
 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with 6-hydroxy-16-methoxy-11-methyl-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(16),3,5,7,9,13(17),14-heptaen-12-one | | Descriptor: | 6-hydroxy-16-methoxy-11-methyl-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(16),3,5,7,9,13(17),14-heptaen-12-one, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Lee, B.I, Park, T.H. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | Synthesis and Structure-Activity Relationships of Aristoyagonine Derivatives as Brd4 Bromodomain Inhibitors with X-ray Co-Crystal Research.
Molecules, 26, 2021
|
|
5C16
 
 | | Myotubularin-related proetin 1 | | Descriptor: | Myotubularin-related protein 1, PHOSPHATE ION | | Authors: | Lee, B.I, Bong, S.M. | | Deposit date: | 2015-06-13 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal Structure of Human Myotubularin-Related Protein 1 Provides Insight into the Structural Basis of Substrate Specificity
Plos One, 11, 2016
|
|
8ZV8
 
 | |
8ZVJ
 
 | |
1UHE
 
 | | Crystal structure of aspartate decarboxylase, isoaspargine complex | | Descriptor: | Aspartate 1-decarboxylase alpha chain, Aspartate 1-decarboxylase beta chain, N~2~-(2-AMINO-1-METHYL-2-OXOETHYLIDENE)ASPARAGINATE | | Authors: | Lee, B.I, Kwon, A.-R, Han, B.W, Suh, S.W. | | Deposit date: | 2003-07-01 | | Release date: | 2004-07-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the schiff base intermediate prior to decarboxylation in the catalytic cycle of aspartate alpha-decarboxylase
J.Mol.Biol., 340, 2004
|
|
1UHD
 
 | | Crystal structure of aspartate decarboxylase, pyruvoly group bound form | | Descriptor: | Aspartate 1-decarboxylase alpha chain, Aspartate 1-decarboxylase beta chain | | Authors: | Lee, B.I, Kwon, A.-R, Han, B.W, Suh, S.W. | | Deposit date: | 2003-07-01 | | Release date: | 2004-07-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the schiff base intermediate prior to decarboxylation in the catalytic cycle of aspartate alpha-decarboxylase
J.Mol.Biol., 340, 2004
|
|
6L4O
 
 | | Crystal structure of API5-FGF2 complex | | Descriptor: | Apoptosis inhibitor 5, Fibroblast growth factor 2 | | Authors: | Lee, B.I, Bong, S.M. | | Deposit date: | 2019-10-18 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Regulation of mRNA export through API5 and nuclear FGF2 interaction.
Nucleic Acids Res., 48, 2020
|
|
1FA2
 
 | | CRYSTAL STRUCTURE OF BETA-AMYLASE FROM SWEET POTATO | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BETA-AMYLASE, alpha-D-glucopyranose-(1-4)-2-deoxy-beta-D-arabino-hexopyranose | | Authors: | Lee, B.I, Cheong, C.G, Suh, S.W. | | Deposit date: | 2000-07-12 | | Release date: | 2000-08-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallization, molecular replacement solution, and refinement of tetrameric beta-amylase from sweet potato.
Proteins, 21, 1995
|
|
5Y5U
 
 | | Crystal structures of spleen tyrosine kinase in complex with a novel inhibitor | | Descriptor: | 4-[(1-methylindazol-5-yl)amino]-2-(4-oxidanylpiperidin-1-yl)-8H-pyrido[4,3-d]pyrimidin-5-one, Tyrosine-protein kinase SYK | | Authors: | Lee, S.J, Lee, B.I. | | Deposit date: | 2017-08-09 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal Structures of Spleen Tyrosine Kinase in Complex with Two Novel 4-Aminopyrido[4,3-d] Pyrimidine Derivative Inhibitors.
Mol. Cells, 41, 2018
|
|
4RNZ
 
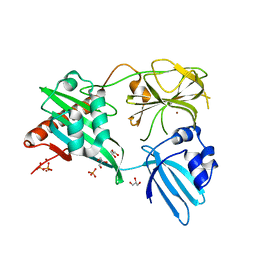 | | Structure of Helicobacter pylori Csd3 from the hexagonal crystal | | Descriptor: | Conserved hypothetical secreted protein, GLYCEROL, NICKEL (II) ION, ... | | Authors: | An, D.R, Kim, H.S, Kim, J, Im, H.N, Yoon, H.J, Yoon, J.Y, Jang, J.Y, Hesek, D, Lee, M, Mobashery, S, Kim, S.-J, Lee, B.I, Suh, S.W. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of Csd3 from Helicobacter pylori, a cell shape-determining metallopeptidase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
