8H0H
 
 | |
8H2D
 
 | |
7R22
 
 | |
1IX1
 
 | | Crystal Structure of P.aeruginosa Peptide deformylase Complexed with Antibiotic Actinonin | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, ACTINONIN, ZINC ION, ... | | Authors: | Kim, H.-W, Yoon, H.-J, Lee, J.Y, Han, B.W, Yang, J.K, Lee, B.I, Ahn, H.J, Lee, H.H, Suh, S.W. | | Deposit date: | 2002-06-07 | | Release date: | 2003-09-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of peptide deformylase from Staphylococcus aureus in complex with actinonin, a naturally occurring antibacterial agent
Proteins, 57, 2004
|
|
4O9W
 
 | |
7YA9
 
 | | Fis1 (Mitochondrial fission 1 protein) | | Descriptor: | BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, Mitochondrial fission 1 protein, ... | | Authors: | Duc, N.M, Bong, S.M, Lee, B.I. | | Deposit date: | 2022-06-27 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insight into the Mitochondria-ER tethering revealed by the crystal structure of Fis1-Bap31 complex
To Be Published
|
|
6JKG
 
 | | The NAD+-free form of human NSDHL | | Descriptor: | Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating | | Authors: | Kim, D, Lee, S.J, Lee, B. | | Deposit date: | 2019-02-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of human NSDHL and development of its novel inhibitor with the potential to suppress EGFR activity.
Cell.Mol.Life Sci., 78, 2021
|
|
4Z9E
 
 | | Alba from Thermoplasma volcanium | | Descriptor: | DNA/RNA-binding protein Alba | | Authors: | Ma, C, Lee, S.J, Pathak, C, Lee, B.J. | | Deposit date: | 2015-04-10 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Alba from Thermoplasma volcanium belongs to alpha-NAT's: An insight into the structural aspects of Tv Alba and its acetylation by Tv Ard1.
Arch.Biochem.Biophys., 590, 2016
|
|
7YKA
 
 | |
3U7M
 
 | | Crystal structures of the Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors | | Descriptor: | N-((2R,4S)-2-butyl-4-(3-(2-fluorophenyl)ureido)-5-methyl-3-oxohexyl)-N-hydroxyformamide, Peptide deformylase, ZINC ION | | Authors: | Lee, S.J, Lee, S.-J, Lee, S.K, Yoon, H.-J, Lee, H.H, Kim, K.K, Lee, B.J, Suh, S.W. | | Deposit date: | 2011-10-14 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3U7K
 
 | | Crystal structures of the Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors | | Descriptor: | (S)-N-(cyclopentylmethyl)-N-(2-(hydroxyamino)-2-oxoethyl)-2-(3-(2-methoxyphenyl)ureido)-3,3-dimethylbutanamide, Peptide deformylase, ZINC ION | | Authors: | Lee, S.J, Lee, S.-J, Lee, S.K, Yoon, H.-J, Lee, H.H, Kim, K.K, Lee, B.J, Suh, S.W. | | Deposit date: | 2011-10-14 | | Release date: | 2012-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3U7N
 
 | | Crystal structures of the Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors | | Descriptor: | N-((2R,4S)-2-butyl-5-methyl-4-(3-(5-methylpyridin-2-yl)ureido)-3-oxohexyl)-N-hydroxyformamide, Peptide deformylase, ZINC ION | | Authors: | Lee, S.J, Lee, S.-J, Lee, S.K, Yoon, H.-J, Lee, H.H, Kim, K.K, Lee, B.J, Suh, S.W. | | Deposit date: | 2011-10-14 | | Release date: | 2012-06-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3U7L
 
 | | Crystal structures of the Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors | | Descriptor: | (S)-N-(cyclopentylmethyl)-2-(3-(3,5-difluorophenyl)ureido)-N-(2-(hydroxyamino)-2-oxoethyl)-3,3-dimethylbutanamide, Peptide deformylase, ZINC ION | | Authors: | Lee, S.J, Lee, S.-J, Lee, S.K, Yoon, H.-J, Lee, H.H, Kim, K.K, Lee, B.J, Suh, S.W. | | Deposit date: | 2011-10-14 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structures of Staphylococcus aureus peptide deformylase in complex with two classes of new inhibitors
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4JX7
 
 | | Crystal structure of Pim1 kinase in complex with inhibitor 2-[(trans-4-aminocyclohexyl)amino]-4-{[3-(trifluoromethyl)phenyl]amino}pyrido[4,3-d]pyrimidin-5(6H)-one | | Descriptor: | 2-[(trans-4-aminocyclohexyl)amino]-4-{[3-(trifluoromethyl)phenyl]amino}pyrido[4,3-d]pyrimidin-5(6H)-one, PIM1 consensus peptide, Serine/threonine-protein kinase pim-1 | | Authors: | Lee, S.J, Han, B.G, Cho, J.W, Choi, J.S, Lee, J.K, Song, H.J, Koh, J.S, Lee, B.I. | | Deposit date: | 2013-03-27 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of pim1 kinase in complex with a pyrido[4,3-d]pyrimidine derivative suggests a unique binding mode.
Plos One, 8, 2013
|
|
4JX3
 
 | | Crystal structure of Pim1 kinase | | Descriptor: | Serine/threonine-protein kinase pim-1 | | Authors: | Lee, S.J, Han, B.G, Cho, J.W, Choi, J.S, Lee, J.K, Song, H.J, Koh, J.S, Lee, B.I. | | Deposit date: | 2013-03-27 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of pim1 kinase in complex with a pyrido[4,3-d]pyrimidine derivative suggests a unique binding mode.
Plos One, 8, 2013
|
|
6JKH
 
 | | The NAD+-bound form of human NSDHL | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating | | Authors: | Kim, D, Lee, S.J, Lee, B. | | Deposit date: | 2019-02-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of human NSDHL and development of its novel inhibitor with the potential to suppress EGFR activity.
Cell.Mol.Life Sci., 78, 2021
|
|
5HS9
 
 | |
5HS7
 
 | |
5HS8
 
 | |
6IFM
 
 | | Crystal structure of DNA bound VapBC from Salmonella typhimurium | | Descriptor: | Antitoxin VapB, DNA backward (27-MER), DNA forward (27-MER), ... | | Authors: | Park, D.W, Lee, B.J. | | Deposit date: | 2018-09-20 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Crystal structure of proteolyzed VapBC and DNA-bound VapBC from Salmonella enterica Typhimurium LT2 and VapC as a putative Ca2+-dependent ribonuclease.
Faseb J., 34, 2020
|
|
5XE2
 
 | | Endoribonuclease from Mycobacterial species | | Descriptor: | Endoribonuclease MazF4 | | Authors: | Ahn, D.-H, Lee, K.-Y, Lee, S.J, Yoon, H.J, Kim, S.-J, Lee, B.-J. | | Deposit date: | 2017-03-31 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural analyses of the MazEF4 toxin-antitoxin pair in Mycobacterium tuberculosis provide evidence for a unique extracellular death factor.
J. Biol. Chem., 292, 2017
|
|
3GIO
 
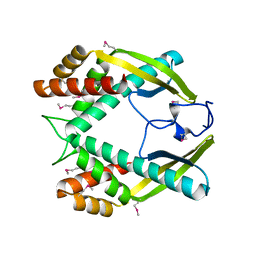 | | Crystal structure of the TNF-alpha inducing protein (Tip alpha) from Helicobacter pylori | | Descriptor: | Putative uncharacterized protein | | Authors: | Jang, J.Y, Yoon, H.J, Yoon, J.Y, Kim, H.S, Lee, S.J, Kim, K.H, Kim, D.J, Han, B.G, Lee, B.I, Jang, S, Suh, S.W. | | Deposit date: | 2009-03-05 | | Release date: | 2009-08-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the TNF-alpha-Inducing Protein (Tipalpha) from Helicobacter pylori: Insights into Its DNA-Binding Activity.
J.Mol.Biol., 2009
|
|
4Y1R
 
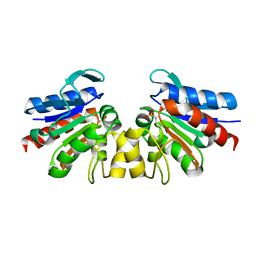 | | SAV1875-cysteinesulfonic acid | | Descriptor: | Uncharacterized protein SAV1875 | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2015-02-08 | | Release date: | 2016-01-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and functional insight into the different oxidation states of SAV1875 from Staphylococcus aureus
Biochem.J., 473, 2016
|
|
5XE3
 
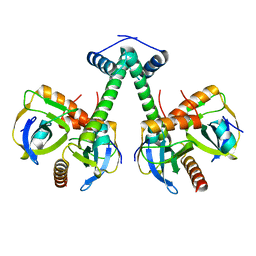 | | Endoribonuclease in complex with its cognate antitoxin from Mycobacterial species | | Descriptor: | Endoribonuclease MazF4, Probable antitoxin MazE4 | | Authors: | Ahn, D.-H, Lee, K.-Y, Lee, S.J, Yoon, H.J, Kim, S.-J, Lee, B.-J. | | Deposit date: | 2017-03-31 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analyses of the MazEF4 toxin-antitoxin pair in Mycobacterium tuberculosis provide evidence for a unique extracellular death factor.
J. Biol. Chem., 292, 2017
|
|
3UI3
 
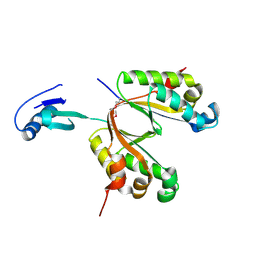 | |
