4OK0
 
 | | Crystal structure of putative nucleotidyltransferase from H. pylori | | Descriptor: | Putative | | Authors: | Yoon, J.Y, Lee, S.J, Lee, B, Yang, J.K, Suh, S.W. | | Deposit date: | 2014-01-21 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of JHP933 from Helicobacter pylori J99 shows two-domain architecture with a DUF1814 family nucleotidyltransferase domain and a helical bundle domain.
Proteins, 82, 2014
|
|
6IFC
 
 | | Crystal structure of VapBC from Salmonella typhimurium | | Descriptor: | Antitoxin VapB, CALCIUM ION, tRNA(fMet)-specific endonuclease VapC | | Authors: | Park, D.W, Lee, B.J. | | Deposit date: | 2018-09-19 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of proteolyzed VapBC and DNA-bound VapBC from Salmonella enterica Typhimurium LT2 and VapC as a putative Ca2+-dependent ribonuclease.
Faseb J., 34, 2020
|
|
2K6P
 
 | | Solution Structure of hypothetical protein, HP1423 | | Descriptor: | Uncharacterized protein HP_1423 | | Authors: | Kim, J, Park, S, Lee, K, Son, W, Sohn, N, Lee, B. | | Deposit date: | 2008-07-15 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hypothetical protein HP1423 (Y1423_HELPY) reveals the presence of alphaL motif related to RNA binding
Proteins, 75, 2009
|
|
3L7U
 
 | | Crystal structure of human NM23-H1 | | Descriptor: | Nucleoside diphosphate kinase A, PHOSPHATE ION | | Authors: | Han, B.G, Min, K, Lee, B.I, Lee, S. | | Deposit date: | 2009-12-29 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Refined Structure of Human NM23-H1 from a Hexagonal Crystal
BULL.KOREAN CHEM.SOC., 31, 2010
|
|
5Y5T
 
 | | Crystal structures of spleen tyrosine kinase in complex with a novel inhibitor | | Descriptor: | 2-[[(1S,2S)-2-azanylcyclohexyl]amino]-4-[(4-methylsulfonylphenyl)amino]-6H-pyrido[4,3-d]pyrimidin-5-one, Tyrosine-protein kinase SYK | | Authors: | Lee, S.J, Lee, B.I. | | Deposit date: | 2017-08-09 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Spleen Tyrosine Kinase in Complex with Two Novel 4-Aminopyrido[4,3-d] Pyrimidine Derivative Inhibitors.
Mol. Cells, 41, 2018
|
|
7E3W
 
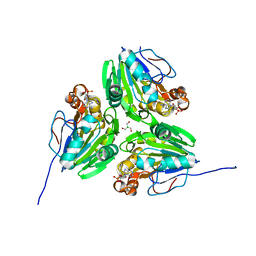 | | Metallo beta-lactamase fold protein (cAMP bound) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Lee, K.-Y, Kim, D.-G, Lee, B.-J. | | Deposit date: | 2021-02-09 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and functional studies of SAV1707 from Staphylococcus aureus elucidate its distinct metal-dependent activity and a crucial residue for catalysis.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7E3V
 
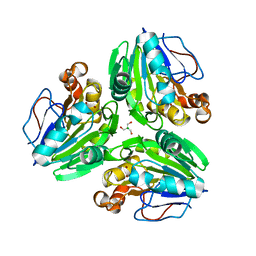 | | Metallo beta-lactamase fold protein (cAMP free) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Lee, K.-Y, Kim, D.-G, Lee, B.-J. | | Deposit date: | 2021-02-09 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and functional studies of SAV1707 from Staphylococcus aureus elucidate its distinct metal-dependent activity and a crucial residue for catalysis.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
4Y1E
 
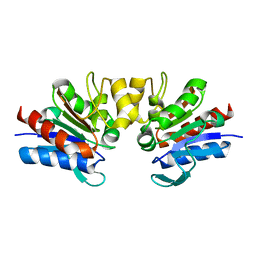 | | SAV1875-C105D | | Descriptor: | Uncharacterized protein SAV1875 | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2015-02-07 | | Release date: | 2016-01-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional insight into the different oxidation states of SAV1875 from Staphylococcus aureus
Biochem.J., 473, 2016
|
|
7W3D
 
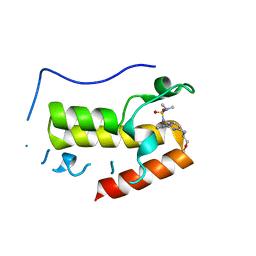 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with N2-(1,2,3-benzotriazol-5-yl)-N3-(dimethylsulfamoyl)-N6-[(2S)-1-methoxypropan-2-yl]pyridine-2,3,6-triamine | | Descriptor: | Bromodomain-containing protein 4, N2-(1,2,3-benzotriazol-5-yl)-N3-(dimethylsulfamoyl)-N6-[(2S)-1-methoxypropan-2-yl]pyridine-2,3,6-triamine | | Authors: | Park, T.H, Lee, B.I. | | Deposit date: | 2021-11-25 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of BET specific bromodomain inhibitors with a novel scaffold.
Bioorg.Med.Chem., 72, 2022
|
|
4Y1F
 
 | | SAV1875-E17D | | Descriptor: | Uncharacterized protein SAV1875 | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2015-02-07 | | Release date: | 2016-01-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional insight into the different oxidation states of SAV1875 from Staphylococcus aureus
Biochem.J., 473, 2016
|
|
4Y0N
 
 | | SAV1875 | | Descriptor: | Uncharacterized protein SAV1875 | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2015-02-06 | | Release date: | 2016-01-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional insight into the different oxidation states of SAV1875 from Staphylococcus aureus
Biochem.J., 473, 2016
|
|
4Y1G
 
 | | SAV1875-E17N | | Descriptor: | Uncharacterized protein SAV1875 | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2015-02-07 | | Release date: | 2016-01-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional insight into the different oxidation states of SAV1875 from Staphylococcus aureus
Biochem.J., 473, 2016
|
|
5K5U
 
 | |
5K63
 
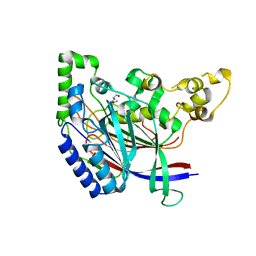 | | Crystal structure of N-terminal amidase C187S | | Descriptor: | ASPARAGINE, GLYCINE, Nta1p | | Authors: | Kim, M.K, Oh, S.-J, Lee, B.-G, Song, H.K. | | Deposit date: | 2016-05-24 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for dual specificity of yeast N-terminal amidase in the N-end rule pathway.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5K66
 
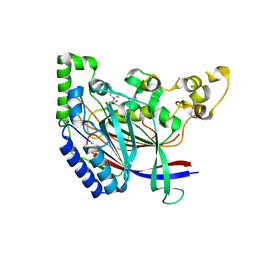 | | Crystal structure of N-terminal amidase with Asn-Glu peptide | | Descriptor: | ASPARAGINE, GLUTAMIC ACID, Nta1p | | Authors: | Kim, M.K, Oh, S.-J, Lee, B.-G, Song, H.K. | | Deposit date: | 2016-05-24 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structural basis for dual specificity of yeast N-terminal amidase in the N-end rule pathway.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5K60
 
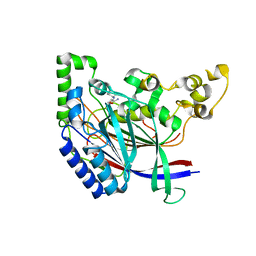 | | Crystal structure of N-terminal amidase with Gln-Val peptide | | Descriptor: | GLUTAMINE, Nta1p, VALINE | | Authors: | Kim, M.K, Oh, S.-J, Lee, B.-G, Song, H.K. | | Deposit date: | 2016-05-24 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for dual specificity of yeast N-terminal amidase in the N-end rule pathway.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5K62
 
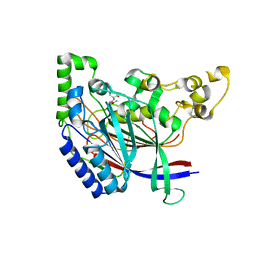 | | Crystal structure of N-terminal amidase C187S | | Descriptor: | ASPARAGINE, Nta1p, VALINE | | Authors: | Kim, M.K, Oh, S.-J, Lee, B.-G, Song, H.K. | | Deposit date: | 2016-05-24 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structural basis for dual specificity of yeast N-terminal amidase in the N-end rule pathway.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5K61
 
 | | Crystal structure of N-terminal amidase with Gln-Gly peptide | | Descriptor: | GLUTAMINE, Nta1p | | Authors: | Kim, M.K, Oh, S.-J, Lee, B.-G, Song, H.K. | | Deposit date: | 2016-05-24 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis for dual specificity of yeast N-terminal amidase in the N-end rule pathway.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5K5V
 
 | |
7YMG
 
 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with 2-({3-ethyl-[1,2,4]triazolo[4,3-b]pyridazin-6-yl}amino)-3-(1H-indol-3-yl)propan-1-ol | | Descriptor: | (2S)-2-[(3-ethyl-[1,2,4]triazolo[4,3-b]pyridazin-6-yl)amino]-3-(1H-indol-3-yl)propan-1-ol, Bromodomain-containing protein 4, FORMIC ACID, ... | | Authors: | Kim, J.H, Lee, B.I. | | Deposit date: | 2022-07-28 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of [1,2,4]triazolo[4,3-b]pyridazine derivatives as BRD4 bromodomain inhibitors and structure-activity relationship study.
Sci Rep, 13, 2023
|
|
7YQ9
 
 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with N-[2-(1H-indol-3-yl)ethyl]-3-(trifluoromethyl)[1,2,4]triazolo[4,3-b]pyridazin-6-amine | | Descriptor: | Bromodomain-containing protein 4, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Kim, J.H, Lee, B.I. | | Deposit date: | 2022-08-05 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of [1,2,4]triazolo[4,3-b]pyridazine derivatives as BRD4 bromodomain inhibitors and structure-activity relationship study.
Sci Rep, 13, 2023
|
|
6YVD
 
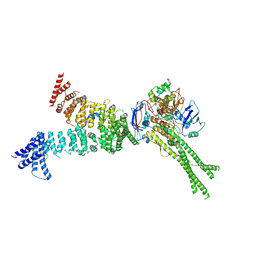 | | Head segment of the S.cerevisiae condensin holocomplex in presence of ATP | | Descriptor: | Condensin complex subunit 2, Condensin complex subunit 3, Structural maintenance of chromosomes protein 2, ... | | Authors: | Merkel, F, Haering, C.H, Hassler, M, Lee, B.G, Lowe, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Cryo-EM structures of holo condensin reveal a subunit flip-flop mechanism.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7VWO
 
 | |
4Q25
 
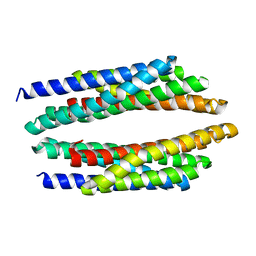 | | Crystal structure of PhoU from Pseudomonas aeruginosa | | Descriptor: | Phosphate-specific transport system accessory protein PhoU homolog | | Authors: | Lee, S.J, Lee, B.-J, Suh, S.W. | | Deposit date: | 2014-04-07 | | Release date: | 2015-02-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of PhoU from Pseudomonas aeruginosa, a negative regulator of the Pho regulon.
J.Struct.Biol., 188, 2014
|
|
5X3T
 
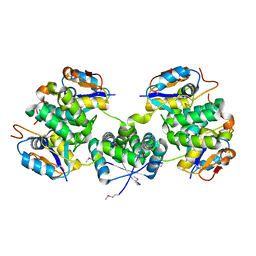 | | VapBC from Mycobacterium tuberculosis | | Descriptor: | Antitoxin VapB26, MAGNESIUM ION, Ribonuclease VapC26 | | Authors: | Kang, S.M, Kim, D.H, Yoon, H.J, Lee, B.J. | | Deposit date: | 2017-02-07 | | Release date: | 2017-06-07 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Functional details of the Mycobacterium tuberculosis VapBC26 toxin-antitoxin system based on a structural study: insights into unique binding and antibiotic peptides.
Nucleic Acids Res., 45, 2017
|
|
