6V5D
 
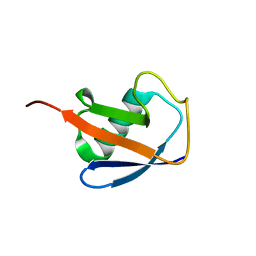 | | EROS3 RDC and NOE Derived Ubiquitin Ensemble | | Descriptor: | Ubiquitin | | Authors: | Lange, O.F, Lakomek, N.A, Smith, C.A, Griesinger, C, de Groot, B.L. | | Deposit date: | 2019-12-04 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Enhancing NMR derived ensembles with kinetics on multiple timescales.
J.Biomol.Nmr, 74, 2020
|
|
2K39
 
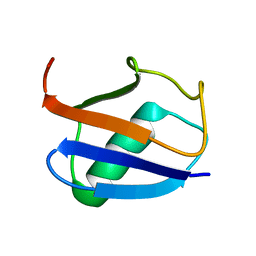 | | Recognition dynamics up to microseconds revealed from RDC derived ubiquitin ensemble in solution | | Descriptor: | Ubiquitin | | Authors: | Lange, O.F, Lakomek, N.A, Fares, C, Schroder, G, Walter, K, Becker, S, Meiler, J, Grubmuller, H, Griesinger, C, de Groot, B.L. | | Deposit date: | 2008-04-25 | | Release date: | 2008-06-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution.
Science, 320, 2008
|
|
2KS6
 
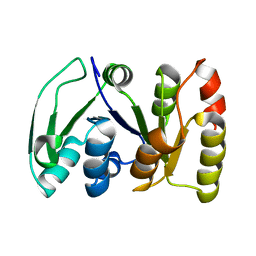 | |
5NPA
 
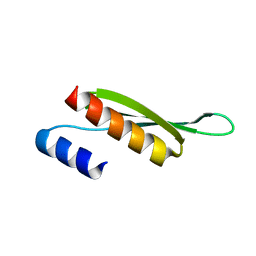 | | Solution structure of Drosophila melanogaster Loquacious dsRBD2 | | Descriptor: | Loquacious | | Authors: | Tants, J.-N, Fesser, S, Kern, T, Stehle, R, Geerlof, A, Wunderlich, C, Boettcher, R, Kunzelmann, S, Lange, O, Kreutz, C, Foerstemann, K, Sattler, M. | | Deposit date: | 2017-04-16 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for asymmetry sensing of siRNAs by the Drosophila Loqs-PD/Dcr-2 complex in RNA interference.
Nucleic Acids Res., 45, 2017
|
|
5NPG
 
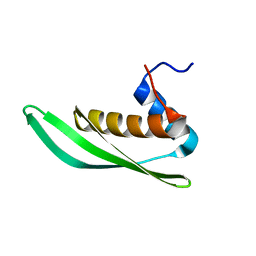 | | Solution structure of Drosophila melanogaster Loquacious dsRBD1 | | Descriptor: | Loquacious, isoform F | | Authors: | Tants, J.-N, Fesser, S, Kern, T, Stehle, R, Geerlof, A, Wunderlich, C, Hartlmuller, C, Boettcher, R, Kunzelmann, S, Lange, O, Kreutz, C, Foerstemann, K, Sattler, M. | | Deposit date: | 2017-04-16 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for asymmetry sensing of siRNAs by the Drosophila Loqs-PD/Dcr-2 complex in RNA interference.
Nucleic Acids Res., 45, 2017
|
|
4QBY
 
 | | yCP in complex with BOC-ALA-ALA-ALA-CHO | | Descriptor: | BOC-ALA-ALA-ALA-CHO, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Arciniega, M, Beck, P, Lange, O, Groll, M, Huber, R. | | Deposit date: | 2014-05-09 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Differential global structural changes in the core particle of yeast and mouse proteasome induced by ligand binding.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2MNG
 
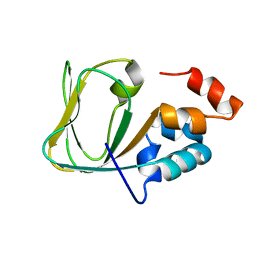 | | Apo Structure of human HCN4 CNBD solved by NMR | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Akimoto, M, Zhang, Z, Boulton, S, Selvaratnam, R, VanSchouwen, B, Gloyd, M, Accili, E.A, Lange, O.F, Melacini, G. | | Deposit date: | 2014-04-03 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A mechanism for the auto-inhibition of hyperpolarization-activated cyclic nucleotide-gated (HCN) channel opening and its relief by cAMP.
J.Biol.Chem., 289, 2014
|
|
2LMD
 
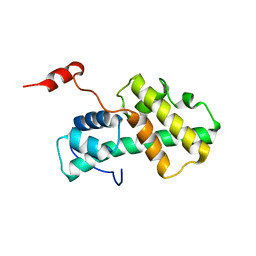 | | Minimal Constraints Solution NMR Structure of Prospero Homeobox protein 1 from Homo sapiens, Northeast Structural Genomics Consortium Target HR4660B | | Descriptor: | Prospero homeobox protein 1 | | Authors: | Rossi, P, Lange, O.A, Lee, H, Maglaqui, M, Janjua, H, Ciccosanti, C, Zhao, L, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-11-29 | | Release date: | 2011-12-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2KW5
 
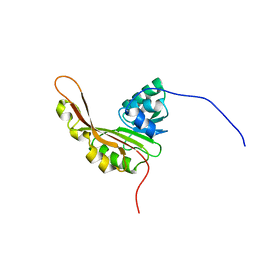 | | Solution NMR Structure of the Slr1183 protein from Synechocystis sp. PCC 6803, Northeast Structural Genomics Consortium Target SgR145 | | Descriptor: | Slr1183 protein | | Authors: | Rossi, P, Forouhar, F, Lee, H, Lange, O, Mao, B, Lemak, A, Maglaqui, M, Belote, R, Ciccosanti, C, Foote, E, Sahdev, S, Acton, T, Xiao, R, Everett, J, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2M89
 
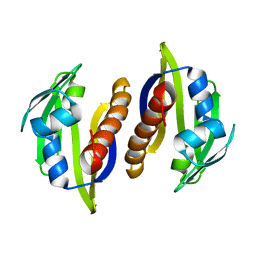 | | Solution structure of the Aha1 dimer from Colwellia psychrerythraea | | Descriptor: | Aha1 domain protein | | Authors: | Rossi, P, Sgourakis, N.G, Shi, L, Liu, G, Barbieri, C.M, Lee, H, Grant, T.D, Luft, J.R, Xiao, R, Acton, T.B, Montelione, G.T, Snell, E.H, Baker, D, Lange, O.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-09 | | Release date: | 2013-09-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | A hybrid NMR/SAXS-based approach for discriminating oligomeric protein interfaces using Rosetta.
Proteins, 83, 2015
|
|
2MV0
 
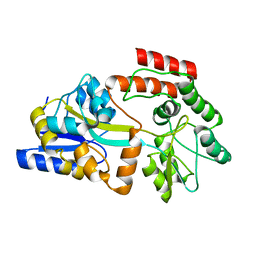 | | Solution NMR Structure of Maltose-binding protein from Escherichia coli, Northeast Structural Genomics Consortium (NESG) Target ER690 | | Descriptor: | Maltose-binding periplasmic protein | | Authors: | Rossi, P, Lange, O.F, Sgourakis, N.G, Song, Y, Lee, H, Aramini, J.M, Ertekin, A, Xiao, R, Acton, T.B, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-09-18 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LOK
 
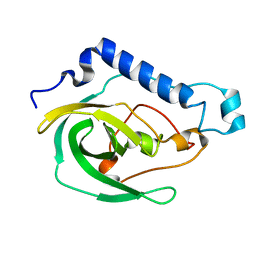 | | Solution NMR Structure of the uncharacterized protein from gene locus VNG_0733H of Halobacterium salinarium, Northeast Structural Genomics Consortium Target HsR50 | | Descriptor: | Uncharacterized protein | | Authors: | Rossi, P, Lange, O.F, Lee, H, Hamilton, K, Ciccosanti, C, Buchwald, W.A, Wang, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-01-24 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LNU
 
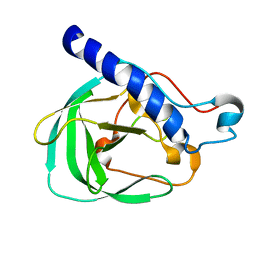 | | Solution NMR Structure of the uncharacterized protein from gene locus rrnAC0354 of Haloarcula marismortui, Northeast Structural Genomics Consortium Target HmR11 | | Descriptor: | Uncharacterized protein | | Authors: | Rossi, P, Liu, G, Lange, O.F, Lee, H, Janjua, H, Ciccosanti, C, Wang, H, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-01-05 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LCB
 
 | | Solution Structure of a Minor and Transiently Formed State of a T4 Lysozyme Mutant | | Descriptor: | Lysozyme | | Authors: | Bouvignies, G, Vallurupalli, P, Hansen, D, Correia, B, Lange, O, Bah, A, Vernon, R.M, Dahlquist, F.W, Baker, D, Kay, L.E. | | Deposit date: | 2011-04-26 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a minor and transiently formed state of a T4 lysozyme mutant.
Nature, 477, 2011
|
|
2LC9
 
 | | Solution Structure of a Minor and Transiently Formed State of a T4 Lysozyme Mutant | | Descriptor: | Lysozyme | | Authors: | Bouvignies, G, Vallurupalli, P, Hansen, D, Correia, B, Lange, O, Bah, A, Vernon, R.M, Dahlquist, F.W, Baker, D, Kay, L.E. | | Deposit date: | 2011-04-26 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a minor and transiently formed state of a T4 lysozyme mutant.
Nature, 477, 2011
|
|
