7N2C
 
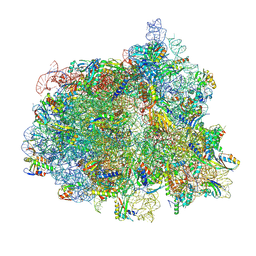 | | Elongating 70S ribosome complex in a fusidic acid-stalled intermediate state of translocation bound to EF-G(GDP) (INT2) | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-28 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
7N31
 
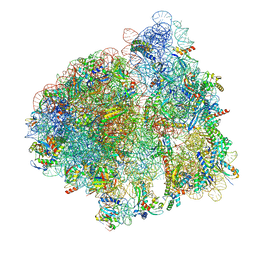 | | Elongating 70S ribosome complex in a post-translocation (POST) conformation | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-31 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
3D0J
 
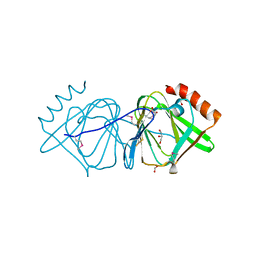 | | Crystal structure of conserved protein of unknown function CA_C3497 from Clostridium acetobutylicum ATCC 824 | | Descriptor: | FORMIC ACID, GLYCEROL, Uncharacterized protein CA_C3497 | | Authors: | Kim, Y, Bigelow, L, Clancy, S, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-01 | | Release date: | 2008-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal Structure of Conserved Protein of Unknown Function CA_C3497 from Clostridium acetobutylicum ATCC 824.
To be Published
|
|
7N2U
 
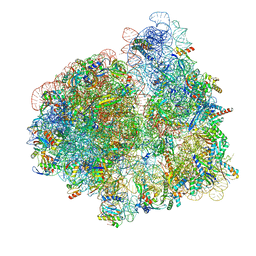 | | Elongating 70S ribosome complex in a hybrid-H1 pre-translocation (PRE-H1) conformation | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Holm, M, Schacherl, M, Natchiar, K.S, Altman, R.B, Spahn, C.M.T, Myasnikov, A.G, Blanchard, S.C. | | Deposit date: | 2021-05-29 | | Release date: | 2021-07-14 | | Last modified: | 2021-08-11 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis of early translocation events on the ribosome.
Nature, 595, 2021
|
|
3DNX
 
 | |
6V71
 
 | | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica with Nitrate in the Active Site | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, FORMIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Clancy, S, Endres, M, Mulligan, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-12-06 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Metallo Beta Lactamase from Hirschia baltica with Nitrate in the Active Site
To Be Published
|
|
6OBZ
 
 | | Crystal structure of FluA-20 Fab | | Descriptor: | Heavy chain of FluA-20 Fab, Light chain of FluA-20 Fab, Immunoglobulin light chain | | Authors: | Wilson, I.A, Lang, S. | | Deposit date: | 2019-03-21 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A Site of Vulnerability on the Influenza Virus Hemagglutinin Head Domain Trimer Interface.
Cell, 177, 2019
|
|
6SM2
 
 | | Mutant immunoglobulin light chain causing amyloidosis (Pat-1) | | Descriptor: | Pat-1 | | Authors: | Kazman, P, Vielberg, M.-T, Cendales, M.D.P, Hunziger, L, Weber, B, Hegenbart, U, Zacharias, M, Koehler, R, Schoenland, S, Groll, M, Buchner, J. | | Deposit date: | 2019-08-21 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fatal amyloid formation in a patient's antibody light chain is caused by a single point mutation.
Elife, 9, 2020
|
|
1QD9
 
 | | Bacillus subtilis YABJ | | Descriptor: | ACETIC ACID, ETHYL MERCURY ION, MERCURY (II) ION, ... | | Authors: | Smith, J.L, Sinha, S, Rappu, P, Lange, S.C, Mantsala, P, Zalkin, H. | | Deposit date: | 1999-07-09 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Bacillus subtilis YabJ, a purine regulatory protein and member of the highly conserved YjgF family.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1YZF
 
 | | Crystal structure of the lipase/acylhydrolase from Enterococcus faecalis | | Descriptor: | lipase/acylhydrolase | | Authors: | Zhang, R, Hatzos, C, Clancy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-28 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the lipase/acylhydrolase from Enterococcus faecalis
To be Published
|
|
3RX9
 
 | | 3D structure of SciN from an Escherichia coli Patotype | | Descriptor: | Putative uncharacterized protein, SULFATE ION | | Authors: | Felisberto-Rodrigues, C, Durand, E, Aschtgen, M.-S, Blangy, S, Ortiz-Lombardia, M, Douzy, B, Cambillau, C, Cascales, E. | | Deposit date: | 2011-05-10 | | Release date: | 2011-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Towards a Structural Comprehension of Bacterial Type VI Secretion Systems: Characterization of the TssJ-TssM Complex of an Escherichia coli Pathovar.
Plos Pathog., 7, 2011
|
|
7R7P
 
 | | Immature HIV-1 CACTD-SP1 lattice with Bevirimat (BVM) and Inositol hexakisphosphate (IP6) | | Descriptor: | 3alpha-[(3-carboxy-3-methylbutanoyl)oxy]-8alpha,9beta,10alpha,13alpha,17alpha,19beta-lup-20(29)-en-28-oic acid, Gag polyprotein, INOSITOL HEXAKISPHOSPHATE | | Authors: | Sarkar, S, Zadrozny, K.K, Zadorozhnyi, R, Russell, R.W, Quinn, C.M, Kleinpeter, A, Ablan, S, Meshkin, H, Perilla, J.R, Ganser-Pornillos, B.K, Pornillos, O, Freed, E.O, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2021-06-25 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structural basis of HIV-1 maturation inhibitor binding and activity.
Nat Commun, 14, 2023
|
|
7R7Q
 
 | | Immature HIV-1 CACTD-SP1 lattice with Inositol hexakisphosphate (IP6) | | Descriptor: | Gag polyprotein, INOSITOL HEXAKISPHOSPHATE | | Authors: | Sarkar, S, Zadrozny, K.K, Zadorozhnyi, R, Russell, R.W, Quinn, C.M, Kleinpeter, A, Ablan, S, Meshkin, H, Perilla, J.R, Ganser-Pornillos, B.K, Pornillos, O, Freed, E.O, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2021-06-25 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structural basis of HIV-1 maturation inhibitor binding and activity.
Nat Commun, 14, 2023
|
|
1QHH
 
 | | STRUCTURE OF DNA HELICASE WITH ADPNP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PROTEIN (PCRA (SUBUNIT)) | | Authors: | Soultanas, P, Dillingham, M.S, Velankar, S.S, Wigley, D.B. | | Deposit date: | 1999-05-14 | | Release date: | 1999-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA binding mediates conformational changes and metal ion coordination in the active site of PcrA helicase.
J.Mol.Biol., 290, 1999
|
|
1Z67
 
 | | Structure of Homeodomain-like Protein of Unknown Function S4005 from Shigella flexneri | | Descriptor: | SODIUM ION, hypothetical protein S4005 | | Authors: | Osipiuk, J, Maltseva, N, Dementieva, I, Clancy, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-03-21 | | Release date: | 2005-05-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of YidB protein from Shigella flexneri shows a new fold with homeodomain motif.
Proteins, 65, 2006
|
|
6TYO
 
 | | Salmonella Typhi PltB Homopentamer with Neu-5NAc-4OAc-alpha-2-3-Gal-beta-1-4-GlcNAc Glycans | | Descriptor: | 4-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Pertussis-like toxin subunit | | Authors: | Nguyen, T, Milano, S.K, Yang, Y.A, Song, J. | | Deposit date: | 2019-08-09 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The role of 9-O-acetylated glycan receptor moieties in the typhoid toxin binding and intoxication.
Plos Pathog., 16, 2020
|
|
8G5Y
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (IC state) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 28S rRNA, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-14 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8GLP
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (Consensus LSU focused refined structure) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Watson, Z.L, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-03-22 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (1.67 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
8G6J
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (GA state 2) | | Descriptor: | (3R,6R,9S,12S,15S,18S,20R,24aR)-6-[(2S)-butan-2-yl]-3,12-bis[(1R)-1-hydroxy-2-methylpropyl]-8,9,11,17,18-pentamethyl-15-[(2S)-2-methylbutyl]hexadecahydropyrido[1,2-a][1,4,7,10,13,16,19]heptaazacyclohenicosine-1,4,7,10,13,16,19(21H)-heptone, (3beta)-O~3~-[(2R)-2,6-dihydroxy-2-(2-methoxy-2-oxoethyl)-6-methylheptanoyl]cephalotaxine, 1,4-DIAMINOBUTANE, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-15 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
4J4Z
 
 | | Crystal structure of the improved variant of the evolved serine hydrolase, OSH55.4_H1.2, bond with sulfate ion in the active site, Northeast Structural Genomics Consortium (NESG) Target OR301 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, Designed serine hydrolase variant OSH55.4_H1.2, ... | | Authors: | Kuzin, A.P, Lew, S, Rajagopalan, S, Maglaqui, M, Xiao, R, Lee, D, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-02-07 | | Release date: | 2013-03-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structure of the improved variant of the evolved serine hydrolase, OSH55.4_H1.2, bond with sulfate ion in the active site, Northeast Structural Genomics Consortium (NESG) Target OR301
To be Published
|
|
6SM1
 
 | | Wild type immunoglobulin light chain (WT-1) | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Immunoglobulin lambda variable 2-14, ... | | Authors: | Kazman, P, Vielberg, M.-T, Cendales, M.D.P, Hunziger, L, Weber, B, Hegenbart, U, Zacharias, M, Koehler, R, Schoenland, S, Groll, M, Buchner, J. | | Deposit date: | 2019-08-21 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Fatal amyloid formation in a patient's antibody light chain is caused by a single point mutation.
Elife, 9, 2020
|
|
8QDI
 
 | | compound 1b bound KMT9 crystal structure | | Descriptor: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[2-[(2~{R})-pyrrolidin-2-yl]ethyl]amino]-2-azanyl-butanoic acid, Methyltransferase N6AMT1, Multifunctional methyltransferase subunit TRM112-like protein | | Authors: | Sheng, W, Eric, M, Roland, S. | | Deposit date: | 2023-08-29 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.467 Å) | | Cite: | compound 1a bound KMT9 crystal structure
To Be Published
|
|
6PL5
 
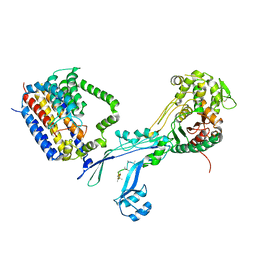 | | Structural coordination of polymerization and crosslinking by a peptidoglycan synthase complex | | Descriptor: | Penicillin-binding protein 2/cell division protein FtsI, Peptidoglycan glycosyltransferase RodA, Unknown peptide | | Authors: | Sjodt, M, Rohs, P.D.A, Erlandson, S.C, Zheng, S, Rudner, D.Z, Bernhardt, T.G, Kruse, A.C. | | Deposit date: | 2019-06-30 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural coordination of polymerization and crosslinking by a SEDS-bPBP peptidoglycan synthase complex.
Nat Microbiol, 5, 2020
|
|
3FIX
 
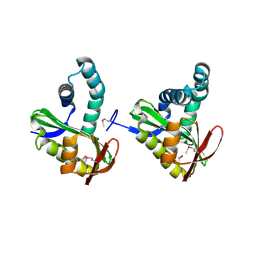 | | Crystal structure of a putative n-acetyltransferase (ta0374) from thermoplasma acidophilum | | Descriptor: | 1,2-ETHANEDIOL, N-ACETYLTRANSFERASE | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-12 | | Release date: | 2009-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the novel PaiA N-acetyltransferase from Thermoplasma acidophilum involved in the negative control of sporulation and degradative enzyme production.
Proteins, 79, 2011
|
|
3M0Z
 
 | | Crystal structure of putative aldolase from Klebsiella pneumoniae. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Chang, C, Rakowski, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-03 | | Release date: | 2010-03-31 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of putative aldolase from Klebsiella pneumoniae.
To be Published
|
|
