4Y0H
 
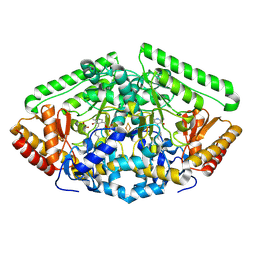 | | Gamma-aminobutyric acid aminotransferase inactivated by (1S,3S)-3-amino-4-difluoromethylenyl-1-cyclopentanoic acid (CPP-115) | | Descriptor: | 4-aminobutyrate aminotransferase, mitochondrial, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Rui, W, Ruslan, S, Hyunbeom, L, Emma, H.D, Jose, I.J, Neil, K, Richard, B.S, Dali, L. | | Deposit date: | 2015-02-06 | | Release date: | 2015-03-11 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Mechanism of inactivation of gamma-aminobutyric acid aminotransferase by (1S,3S)-3-amino-4-difluoromethylenyl-1-cyclopentanoic acid (CPP-115)
J. Am. Chem. Soc., 2015
|
|
3J0O
 
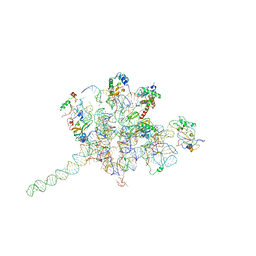 | | Core of mammalian 80S pre-ribosome in complex with tRNAs fitted to a 9A cryo-EM map: classic PRE state 2 | | Descriptor: | 40S ribosomal RNA fragment, 60S ribosomal RNA fragment, Ribosomal protein L10a, ... | | Authors: | Budkevich, T, Giesebrecht, J, Altman, R, Munro, J, Mielke, T, Nierhaus, K, Blanchard, S, Spahn, C.M. | | Deposit date: | 2011-10-05 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structure and dynamics of the Mammalian ribosomal pretranslocation complex.
Mol.Cell, 44, 2011
|
|
2KFH
 
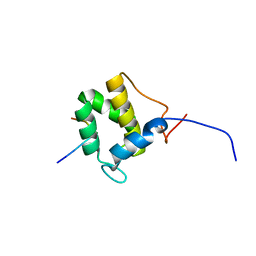 | | Structure of the C-terminal domain of EHD1 with FNYESTGPFTAK | | Descriptor: | CALCIUM ION, EH domain-containing protein 1, Rab11-FIP2 GPF peptide FNYESTGPFTAK | | Authors: | Kieken, F, Jovic, M, Tonelli, M, Naslavsky, N, Caplan, S, Sorgen, P. | | Deposit date: | 2009-02-20 | | Release date: | 2009-12-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the interaction of proteins containing NPF, DPF, and GPF motifs with the C-terminal EH-domain of EHD1.
Protein Sci., 18, 2009
|
|
3J0Q
 
 | | Core of mammalian 80S pre-ribosome in complex with tRNAs fitted to a 10.6A cryo-em map: rotated PRE state 2 | | Descriptor: | 40S ribosomal RNA fragment, 60S ribosomal RNA fragment, Ribosomal protein L10, ... | | Authors: | Budkevich, T, Giesebrecht, J, Altman, R, Munro, J, Mielke, T, Nierhaus, K, Blanchard, S, Spahn, C.M. | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (10.6 Å) | | Cite: | Structure and dynamics of the Mammalian ribosomal pretranslocation complex.
Mol.Cell, 44, 2011
|
|
3J0P
 
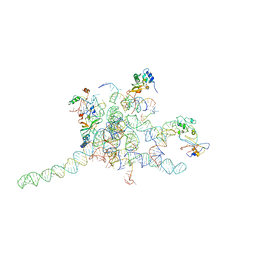 | | Core of mammalian 80S pre-ribosome in complex with tRNAs fitted to a 10.6A cryo-em map: rotated PRE state 1 | | Descriptor: | 40S ribosomal RNA fragment, 60S ribosomal RNA fragment, Ribosomal protein L10a, ... | | Authors: | Budkevich, T, Giesebrecht, J, Altman, R, Munro, J, Mielke, T, Nierhaus, K, Blanchard, S, Spahn, C.M. | | Deposit date: | 2011-10-06 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (10.6 Å) | | Cite: | Structure and dynamics of the Mammalian ribosomal pretranslocation complex.
Mol.Cell, 44, 2011
|
|
3F6N
 
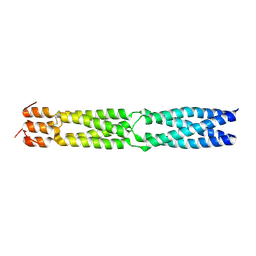 | |
6UK5
 
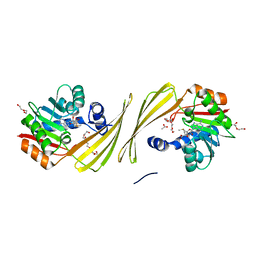 | | Structure of SAM bound CalS10, an amino pentose methyltransferase from Micromonospora echinaspora involved in calicheamicin biosynthesis | | Descriptor: | ACETATE ION, CalS10, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Alvarado, S.K, Miller, M.D, Xu, W, Wang, Z, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-10-04 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of SAM bound CalS10, an amino pentose methyltransferase from Micromonospora echinaspora involved in calicheamicin biosynthesis
To Be Published
|
|
3JRK
 
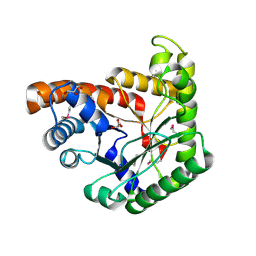 | | A putative tagatose 1,6-diphosphate aldolase from Streptococcus pyogenes | | Descriptor: | GLYCEROL, Tagatose 1,6-diphosphate aldolase 2 | | Authors: | Fan, Y, Volkart, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-08 | | Release date: | 2009-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The crystal structure of a putative tagatose 1,6-diphosphate aldolase from Streptococcus pyogenes
To be Published
|
|
4YBB
 
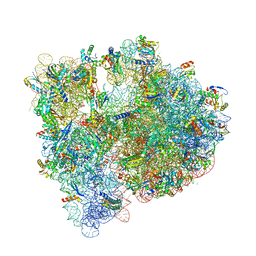 | | High-resolution structure of the Escherichia coli ribosome | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Noeske, J, Wasserman, M.R, Terry, D.S, Altman, R.B, Blanchard, S.C, Cate, J.H.D. | | Deposit date: | 2015-02-18 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution structure of the Escherichia coli ribosome.
Nat.Struct.Mol.Biol., 22, 2015
|
|
7R4U
 
 | | Apoform of FtrA/P19 from Rubrivivax gelatinosus | | Descriptor: | FtrA-P19, GLYCEROL, SODIUM ION, ... | | Authors: | Morera, S, Vigouroux, A, Plancqueel, S. | | Deposit date: | 2022-02-09 | | Release date: | 2022-05-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | New insights into the mechanism of iron transport through the bacterial Ftr system present in pathogens.
Febs J., 289, 2022
|
|
4L99
 
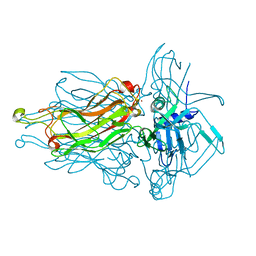 | | Structure of the RBP from lactococcal phage 1358 in complex with glycerol | | Descriptor: | GLYCEROL, Receptor Binding Protein, ZINC ION | | Authors: | Farenc, C, Spinelli, S, Bebeacua, C, Tremblay, D, Orlov, I, Blangy, S, Klaholz, B.P, Moineau, S, Cambillau, C. | | Deposit date: | 2013-06-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Virulent Siphophage CyoEM Structure and Host Recognition and Infection Mechanism
To be Published
|
|
2KFF
 
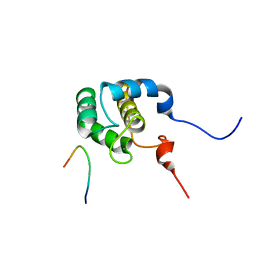 | | Structure of the C-terminal domain of EHD1 with FNYESTNPFTAK | | Descriptor: | CALCIUM ION, EH domain-containing protein 1, Rab11-FIP2 NPF peptide FNYESTNPFTAK | | Authors: | Kieken, F, Jovic, M, Tonelli, M, Naslavsky, N, Caplan, S, Sorgen, P. | | Deposit date: | 2009-02-20 | | Release date: | 2009-12-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the interaction of proteins containing NPF, DPF, and GPF motifs with the C-terminal EH-domain of EHD1.
Protein Sci., 18, 2009
|
|
2OCZ
 
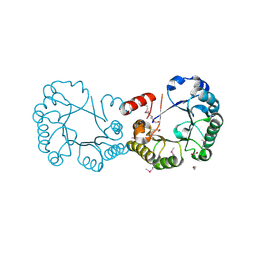 | | The Structure of a Putative 3-Dehydroquinate Dehydratase from Streptococcus pyogenes. | | Descriptor: | 1,2-ETHANEDIOL, 3-dehydroquinate dehydratase, MAGNESIUM ION | | Authors: | Cuff, M.E, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-21 | | Release date: | 2007-01-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of a Putative 3-Dehydroquinate Dehydratase from Streptococcus pyogenes.
TO BE PUBLISHED
|
|
2P0T
 
 | | Structural Genomics, the crystal structure of a conserved putative protein from Pseudomonas syringae pv. tomato str. DC3000 | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, UPF0307 protein PSPTO_4464 | | Authors: | Tan, K, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-03-01 | | Release date: | 2007-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The crystal structure of a conserved putative protein from Pseudomonas syringae pv. tomato str. DC3000
To be Published
|
|
3N7A
 
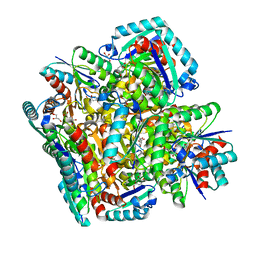 | | Crystal structure of 3-dehydroquinate dehydratase from Mycobacterium tuberculosis in complex with inhibitor 2 | | Descriptor: | 2,3 -ANHYDRO-QUINIC ACID, 3-dehydroquinate dehydratase, GLYCEROL | | Authors: | Dias, M.V.B, Snee, W.C, Bromfield, K.M, Payne, R, Palaninathan, S.K, Ciulli, A, Howard, N.I, Abell, C, Sacchettini, J.C, Blundell, T.L. | | Deposit date: | 2010-05-26 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
3N8N
 
 | | Crystal structure of 3-dehydroquinate dehydratase from Mycobacterium tuberculosis in complex with inhibitor 6 | | Descriptor: | (1R,4R,5R)-3-(tert-butylcarbamoyl)-1,4,5-trihydroxycyclohex-2-ene-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Dias, M.V.B, Snee, W.C, Bromfield, K.M, Payne, R, Palaninathan, S.K, Ciulli, A, Howard, N.I, Abell, C, Sacchettini, J.C, Blundell, T.L. | | Deposit date: | 2010-05-28 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
4WOT
 
 | | ROCK2 IN COMPLEX WITH 1426382-07-1 | | Descriptor: | Rho-associated protein kinase 2, methyl 3-[({2'-(aminomethyl)-5'-[(3-fluoropyridin-4-yl)carbamoyl]biphenyl-3-yl}carbonyl)amino]-4-fluorobenzoate | | Authors: | Augustin, M, Krapp, S, Boland, S, Defert, O, Bourin, A, Alen, J, Leysen, D. | | Deposit date: | 2014-10-16 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Design, synthesis, and biological evaluation of novel, highly active soft ROCK inhibitors.
J. Med. Chem., 58, 2015
|
|
2PKK
 
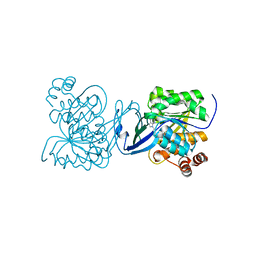 | | Crystal structure of M tuberculosis Adenosine Kinase complexed with 2-fluro adenosine | | Descriptor: | 2-(6-AMINO-2-FLUORO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, Adenosine kinase | | Authors: | Reddy, M.C.M, Palaninathan, S.K, Shetty, N.D, Owen, J.L, Watson, M.D, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-04-17 | | Release date: | 2007-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | High resolution crystal structures of Mycobacterium tuberculosis adenosine kinase: insights into the mechanism and specificity of this novel prokaryotic enzyme
J.Biol.Chem., 282, 2007
|
|
4WOI
 
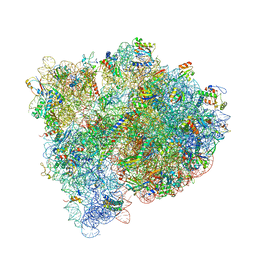 | | 4,5-linked aminoglycoside antibiotics regulate the bacterial ribosome by targeting dynamic conformational processes within intersubunit bridge B2 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Pulk, A, Cate, J.H.D, Blanchard, S, Wasserman, M, Altman, R, Zhou, Z, Zinder, J, Green, K, Garneau-Tsodikova, S. | | Deposit date: | 2014-10-15 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Chemically related 4,5-linked aminoglycoside antibiotics drive subunit rotation in opposite directions.
Nat Commun, 6, 2015
|
|
1UWF
 
 | | 1.7 A resolution structure of the receptor binding domain of the FimH adhesin from uropathogenic E. coli | | Descriptor: | FIMH PROTEIN, GLYCEROL, butyl alpha-D-mannopyranoside | | Authors: | Bouckaert, J, Berglund, J, Genst, E.D, Cools, L, Hung, C.-S, Wuhrer, M, Zavialov, A, Langermann, S, Hultgren, S, Wyns, L, Oscarson, S, Knight, S.D, De Greve, H. | | Deposit date: | 2004-02-05 | | Release date: | 2005-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Receptor Binding Studies Disclose a Novel Class of High-Affinity Inhibitors of the Escherichia Coli Fimh Adhesin.
Mol.Microbiol., 55, 2005
|
|
4KYB
 
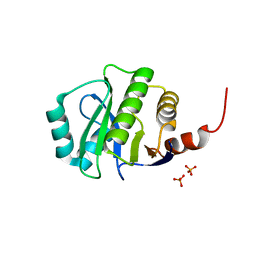 | | Crystal Structure of de novo designed serine hydrolase OSH55.14_E3, Northeast Structural Genomics Consortium Target OR342 | | Descriptor: | Designed Protein OR342, PHOSPHATE ION | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Mao, L, Xiao, R, Lee, D, Raja, S, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-28 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.909 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR342
To be Published
|
|
4L97
 
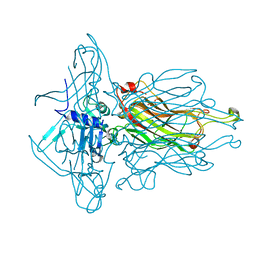 | | Structure of the RBP of lactococcal phage 1358 in complex with glucose-1-phosphate | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, Receptor Binding Protein | | Authors: | Farenc, C, Spinelli, S, Bebeacua, C, Tremblay, D, Orlov, I, Blangy, S, Klaholz, B.P, Moineau, S, Cambillau, C. | | Deposit date: | 2013-06-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | A Virulent Siphophage CyoEM Structure and Host Recognition and Infection Mechanism
To be Published
|
|
3FXW
 
 | | High resolution crystal structure of mitogen-activated protein kinase-activated protein kinase 3/inhibitor 2 complex | | Descriptor: | 2-[2-(2-FLUOROPHENYL)PYRIDIN-4-YL]-1,5,6,7-TETRAHYDRO-4H-PYRROLO[3,2-C]PYRIDIN-4-ONE, MAP kinase-activated protein kinase 3 | | Authors: | Cheng, R.K.Y, Barker, J, Palan, S, Felicetti, B, Whittaker, M, Hesterkamp, T. | | Deposit date: | 2009-01-21 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High resolution crystal structure of mitogen-activated protein kinase-activated protein kinase 3/inhibitor 2 complex
To be Published
|
|
8QDG
 
 | | compound 1a bound KMT9 crystal structure | | Descriptor: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[(3~{S})-pyrrolidin-3-yl]amino]-2-azanyl-butanoic acid, Methyltransferase N6AMT1, Multifunctional methyltransferase subunit TRM112-like protein | | Authors: | Sheng, W, Eric, M, Roland, S. | | Deposit date: | 2023-08-29 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.392 Å) | | Cite: | compound 1a bound KMT9 crystal structure
To Be Published
|
|
2O3C
 
 | | Crystal structure of zebrafish Ape | | Descriptor: | APEX nuclease 1, LEAD (II) ION | | Authors: | Georgiadis, M.M, Gaur, R.K, Delaplane, S, Svenson, J. | | Deposit date: | 2006-12-01 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease
Mutat.Res., 643, 2008
|
|
