4BMJ
 
 | | Structure of the UBZ1and2 tandem of the ubiquitin-binding adaptor protein TAX1BP1 | | Descriptor: | CHLORIDE ION, TAX1-BINDING PROTEIN 1, ZINC ION | | Authors: | Ceregido, M.A, Spinola-Amilibia, M, Buts, L, Rivera, J, Bravo, J, van Nuland, N.A.J. | | Deposit date: | 2013-05-09 | | Release date: | 2013-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Structure of Tax1BP1 Ubz1 + 2 Provides Insight Into Target Specificity and Adaptability
J.Mol.Biol., 426, 2014
|
|
4U17
 
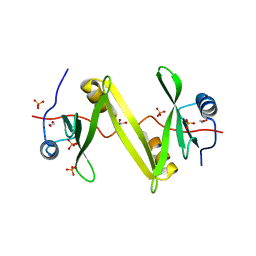 | | Swapped dimer of the human Fyn-SH2 domain | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Tyrosine-protein kinase Fyn | | Authors: | Garcia-Pino, A, Huculeci, R, Lenaerts, T, van Nuland, N.A.J. | | Deposit date: | 2014-07-15 | | Release date: | 2015-07-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Swapped dimer of the human Fyn-SH2 domain
To Be Published
|
|
6BBT
 
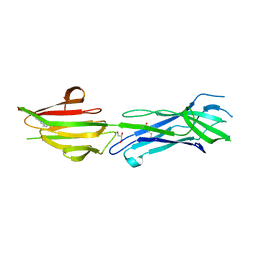 | | Structure of the major pilin protein (T-13) from Streptococcus pyogenes serotype GAS131465 | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Young, P.G, Baker, E.N, Moreland, N.J. | | Deposit date: | 2017-10-19 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Group AStreptococcusT Antigens Have a Highly Conserved Structure Concealed under a Heterogeneous Surface That Has Implications for Vaccine Design.
Infect.Immun., 87, 2019
|
|
1TYO
 
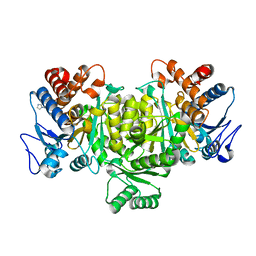 | | Isocitrate Dehydrogenase from the hyperthermophile Aeropyrum pernix in complex with etheno-NADP | | Descriptor: | ETHENO-NADP, isocitrate dehydrogenase | | Authors: | Karlstrom, M, Stokke, R, Steen, I.H, Birkeland, N, Ladenstein, R. | | Deposit date: | 2004-07-08 | | Release date: | 2005-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Isocitrate dehydrogenase from the hyperthermophile Aeropyrum pernix: X-ray structure analysis of a ternary enzyme-substrate complex and thermal stability
J.Mol.Biol., 345, 2005
|
|
2X0J
 
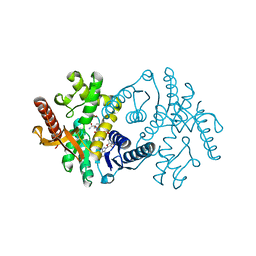 | | 2.8 A RESOLUTION STRUCTURE OF MALATE DEHYDROGENASE FROM ARCHAEOGLOBUS FULGIDUS IN COMPLEX WITH ETHENO-NAD | | Descriptor: | ETHENO-NAD, MALATE DEHYDROGENASE, SULFATE ION | | Authors: | Irimia, A, Madern, D, Zaccai, G, Vellieux, F.M, Karshikoff, A, Tibbelin, G, Ladenstein, R, Lien, T, Birkeland, N.K. | | Deposit date: | 2009-12-14 | | Release date: | 2009-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.786 Å) | | Cite: | The 2.9A Resolution Crystal Structure of Malate Dehydrogenase from Archaeoglobus Fulgidus: Mechanisms of Oligomerisation and Thermal Stabilisation.
J.Mol.Biol., 335, 2004
|
|
2ION
 
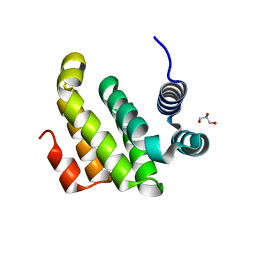 | |
6UNV
 
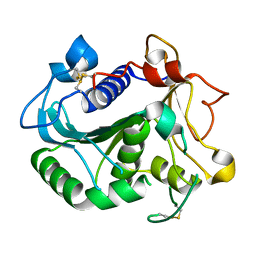 | |
3ANU
 
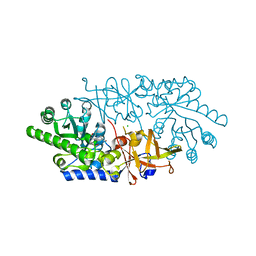 | | Crystal structure of D-serine dehydratase from chicken kidney | | Descriptor: | CHLORIDE ION, D-serine dehydratase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Tanaka, H, Senda, M, Venugopalan, N, Yamamoto, A, Senda, T, Ishida, T, Horiike, K. | | Deposit date: | 2010-09-09 | | Release date: | 2011-06-15 | | Last modified: | 2013-08-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a zinc-dependent D-serine dehydratase from chicken kidney
J.Biol.Chem., 286, 2011
|
|
6BBW
 
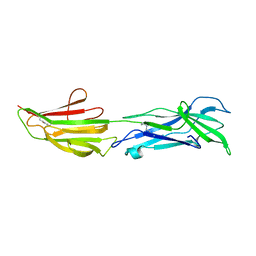 | |
2NSZ
 
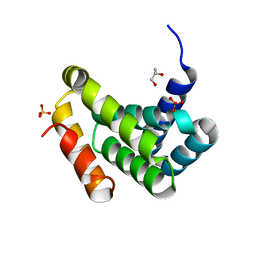 | |
2XE4
 
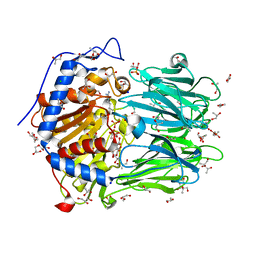 | | Structure of Oligopeptidase B from Leishmania major | | Descriptor: | ANTIPAIN, CHLORIDE ION, GLYCEROL, ... | | Authors: | McLuskey, K, Paterson, N.G, Bland, N.D, Mottram, J.C, Isaacs, N.W. | | Deposit date: | 2010-05-11 | | Release date: | 2010-10-06 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Leishmania Major Oligopeptidase B Gives Insight Into the Enzymatic Properties of a Trypanosomatid Virulence Factor.
J.Biol.Chem., 285, 2010
|
|
4D4U
 
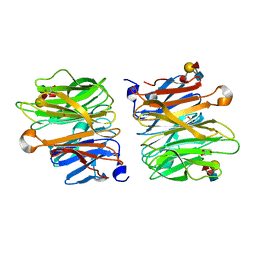 | | Crystal Structure of Fucose binding lectin from Aspergillus Fumigatus (AFL) in complex with LewisY tetrasaccharide. | | Descriptor: | FUCOSE-SPECIFIC LECTIN FLEA, GLYCEROL, alpha-L-fucopyranose, ... | | Authors: | Houser, J, Komarek, J, Kostlanova, N, Lahmann, M, Cioci, G, Varrot, A, Imberty, A, Wimmerova, M. | | Deposit date: | 2014-10-31 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Insights Into Aspergillus Fumigatus Lectin Specificity: Afl Binding Sites are Functionally Non-Equivalent.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3SDG
 
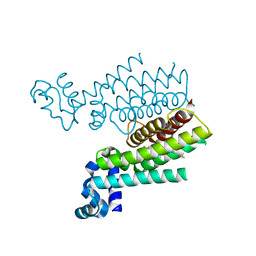 | | Ethionamide Boosters Part 2: Combining Bioisosteric Replacement and Structure-Based Drug Design to Solve Pharmacokinetic Issues in a Series of Potent 1,2,4-Oxadiazole EthR Inhibitors. | | Descriptor: | 4,4,4-trifluoro-1-{4-[3-(1,3-thiazol-2-yl)-1,2,4-oxadiazol-5-yl]piperidin-1-yl}butan-1-one, HTH-type transcriptional regulator EthR | | Authors: | Flipo, M, Desroses, M, Lecat-Guillet, N, Villemagne, B, Blondiaux, N, Leroux, F, Piveteau, C, Mathys, V, Flament, M.P, Siepmann, J, Villeret, V, Wohlkonig, A, Wintjens, R, Soror, S.H, Christophe, T, Jeon, H.K, Locht, C, Brodin, P, D prez, B, Baulard, A.R, Willand, N. | | Deposit date: | 2011-06-09 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Ethionamide Boosters. 2. Combining Bioisosteric Replacement and Structure-Based Drug Design To Solve Pharmacokinetic Issues in a Series of Potent 1,2,4-Oxadiazole EthR Inhibitors.
J.Med.Chem., 55, 2012
|
|
3SFI
 
 | | Ethionamide Boosters Part 2: Combining Bioisosteric Replacement and Structure-Based Drug Design to Solve Pharmacokinetic Issues in a Series of Potent 1,2,4-Oxadiazole EthR Inhibitors. | | Descriptor: | 5,5,5-trifluoro-1-{4-[3-(1,3-thiazol-2-yl)-1,2,4-oxadiazol-5-yl]piperidin-1-yl}pentan-1-one, Transcriptional regulatory repressor protein (TETR-family) | | Authors: | Flipo, M, Desroses, M, Lecat-Guillet, N, Villemagne, B, Blondiaux, N, Leroux, F, Piveteau, C, Mathys, V, Flament, M.P, Siepmann, J, Villeret, V, Wohlkonig, A, Wintjens, R, Soror, S.H, Christophe, T, Jeon, H.K, Locht, C, Brodin, P, D prez, B, Baulard, A.R, Willand, N. | | Deposit date: | 2011-06-13 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Ethionamide Boosters. 2. Combining Bioisosteric Replacement and Structure-Based Drug Design To Solve Pharmacokinetic Issues in a Series of Potent 1,2,4-Oxadiazole EthR Inhibitors.
J.Med.Chem., 55, 2012
|
|
3B4Y
 
 | | FGD1 (Rv0407) from Mycobacterium tuberculosis | | Descriptor: | CITRATE ANION, COENZYME F420, PROBABLE F420-DEPENDENT GLUCOSE-6-PHOSPHATE DEHYDROGENASE FGD1 | | Authors: | Bashiri, G, Squire, C.J, Moreland, N.M, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-10-25 | | Release date: | 2008-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of F420-dependent glucose-6-phosphate dehydrogenase FGD1 involved in the activation of the anti-tuberculosis drug candidate PA-824 reveal the basis of coenzyme and substrate binding
J.Biol.Chem., 283, 2008
|
|
1ODB
 
 | | THE CRYSTAL STRUCTURE OF HUMAN S100A12 - COPPER COMPLEX | | Descriptor: | CALCIUM ION, CALGRANULIN C, COPPER (II) ION | | Authors: | Moroz, O.V, Antson, A.A, Grist, S.J, Maitland, N.J, Dodson, G.G, Wilson, K.S, Lukanidin, E.M, Bronstein, I.B. | | Deposit date: | 2003-02-15 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of the Human S100A12-Copper Complex: Implications for Host-Parasite Defence
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1W9N
 
 | | Isolation and characterization of epilancin 15X, a novel antibiotic from a clinical strain of Staphylococcus epidermidis | | Descriptor: | EPILANCIN 15X | | Authors: | Ekkelenkamp, M, Hanssen, M.G.M, Hsu, S.-T.D, de Jong, A, Milatovic, D, Verhoef, J, van Nuland, N.A.J. | | Deposit date: | 2004-10-14 | | Release date: | 2005-04-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Isolation and structural characterization of epilancin 15X, a novel lantibiotic from a clinical strain of Staphylococcus epidermidis.
FEBS Lett., 579, 2005
|
|
4DVZ
 
 | | Crystal structure of the Helicobacter pylori CagA oncoprotein | | Descriptor: | Cytotoxicity-associated immunodominant antigen | | Authors: | Hayashi, T, Senda, M, Morohashi, H, Higashi, H, Horio, M, Kashiba, Y, Nagase, L, Sasaya, D, Shimizu, T, Venugopalan, N, Kumeta, H, Noda, N, Inagaki, F, Senda, T, Hatakeyama, M. | | Deposit date: | 2012-02-23 | | Release date: | 2012-07-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Tertiary structure-function analysis reveals the pathogenic signaling potentiation mechanism of Helicobacter pylori oncogenic effector CagA
Cell Host Microbe, 12, 2012
|
|
4DW6
 
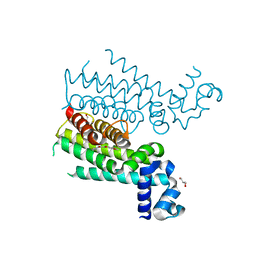 | | Novel N-phenyl-phenoxyacetamide derivatives as potential EthR inhibitors and ethionamide boosters. Discovery and optimization using High-Throughput Synthesis. | | Descriptor: | AMMONIUM ION, GLYCEROL, HTH-type transcriptional regulator EthR, ... | | Authors: | Flipo, M, Willand, N, Lecat-Guillet, N, Hounsou, C, Desroses, M, Leroux, F, Lens, Z, Villeret, V, Wohlkonig, A, Wintjens, R, Christophe, T, Jeon, H.K, Locht, C, Brodin, P, Baulard, A.R, Deprez, B. | | Deposit date: | 2012-02-24 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of novel N-phenylphenoxyacetamide derivatives as EthR inhibitors and ethionamide boosters by combining high-throughput screening and synthesis.
J.Med.Chem., 55, 2012
|
|
4DVY
 
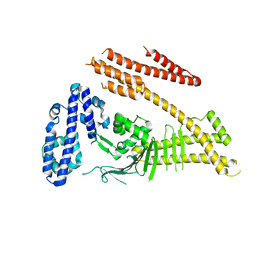 | | Crystal structure of the Helicobacter pylori CagA oncoprotein | | Descriptor: | Cytotoxicity-associated immunodominant antigen | | Authors: | Hayashi, T, Senda, M, Morohashi, H, Higashi, H, Horio, M, Kashiba, Y, Nagase, L, Sasaya, D, Shimizu, T, Venugopalan, N, Kumeta, H, Noda, N, Inagaki, F, Senda, T, Hatakeyama, M. | | Deposit date: | 2012-02-23 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Tertiary Structure-Function Analysis Reveals the Pathogenic Signaling Potentiation Mechanism of Helicobacter pylori Oncogenic Effector CagA
Cell Host Microbe, 12, 2012
|
|
4PHL
 
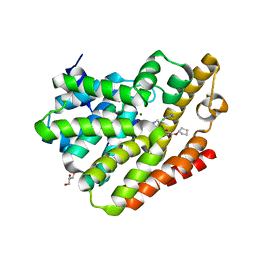 | | TbrPDEB1-inhibitor complex | | Descriptor: | 3-(CYCLOPENTYLOXY)-N-(3,5-DICHLOROPYRIDIN-4-YL)-4-METHOXYBENZAMIDE, Class 1 phosphodiesterase PDEB1, ETHANOL, ... | | Authors: | Choy, M.S, Bland, N, Peti, W, Page, R. | | Deposit date: | 2014-05-06 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | TbrPDEB1-inhibitor complex
To Be Published
|
|
4AGT
 
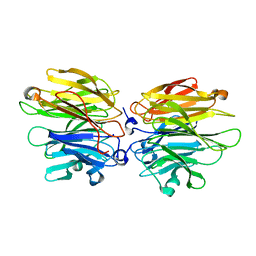 | | Crystal Structure of Fucose binding lectin from Aspergillus Fumigatus (AFL) in complex with Fuc1-6GlcNAc. | | Descriptor: | FUCOSE-SPECIFIC LECTIN FLEA, SODIUM ION, alpha-L-fucopyranose, ... | | Authors: | Houser, J, Komarek, J, Kostlanova, N, Lahmann, M, Cioci, G, Varrot, A, Imberty, A, Wimmerova, M. | | Deposit date: | 2012-01-31 | | Release date: | 2013-02-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights Into Aspergillus Fumigatus Lectin Specificity: Afl Binding Sites are Functionally Non-Equivalent
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5A3J
 
 | | Crystal structure of the chloroplastic gamma-ketol reductase from Arabidopsis thaliana bound to 13-Oxo-9(Z),11(E),15(Z)- octadecatrienoic acid. | | Descriptor: | (13-oxo-9(Z),11(E),15(Z)-octadecatrienoic acid), PUTATIVE QUINONE-OXIDOREDUCTASE HOMOLOG, CHLOROPLASTIC | | Authors: | Mas-y-mas, S, Curien, G, Giustini, C, Rolland, N, Ferrer, J.L, Cobessi, D. | | Deposit date: | 2015-06-01 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.776 Å) | | Cite: | Crystal Structure of the Chloroplastic Oxoene Reductase ceQORH from Arabidopsis thaliana.
Front Plant Sci, 8, 2017
|
|
1T04
 
 | | Three dimensional structure of a humanized anti-IFN-Gamma Fab in C2 space group | | Descriptor: | Huzaf Antibody Heavy Chain, Huzaf Antibody Light Chain | | Authors: | Bourne, P.C, Terzyan, S.S, Cloud, G, Landolfi, N.F, Vasquez, M, Edmundson, A.B. | | Deposit date: | 2004-04-07 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-dimensional structures of a humanized anti-IFN-gamma Fab (HuZAF) in two crystal forms.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1T3F
 
 | | THREE DIMENSIONAL STRUCTURE OF A HUMANIZED ANTI-IFN-GAMMA FAB (HuZAF) IN P21 21 21 SPACE GROUP | | Descriptor: | Huzaf antibody heavy chain, Huzaf antibody light chain | | Authors: | Bourne, P.C, Terzyan, S.S, Cloud, G, Landolfi, N.F, Vasquez, M, Edmundson, A.B. | | Deposit date: | 2004-04-26 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structures of a humanized anti-IFN-gamma Fab (HuZAF) in two crystal forms.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
