7MYL
 
 | |
7MYM
 
 | | Crystal structure of Escherichia coli dihydrofolate reductase in complex with TRIMETHOPRIM and NADPH | | 分子名称: | ARGININE, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Erlandsen, H, Wright, D, Krucinska, J. | | 登録日 | 2021-05-21 | | 公開日 | 2022-06-01 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.04 Å) | | 主引用文献 | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
6PAH
 
 | | HUMAN PHENYLALANINE HYDROXYLASE CATALYTIC DOMAIN DIMER WITH BOUND L-DOPA (3,4-DIHYDROXYPHENYLALANINE) INHIBITOR | | 分子名称: | 3,4-DIHYDROXYPHENYLALANINE, FE (III) ION, PHENYLALANINE 4-MONOOXYGENASE | | 著者 | Erlandsen, H, Flatmark, T, Stevens, R.C. | | 登録日 | 1998-08-20 | | 公開日 | 1999-04-27 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystallographic analysis of the human phenylalanine hydroxylase catalytic domain with bound catechol inhibitors at 2.0 A resolution.
Biochemistry, 37, 1998
|
|
1H15
 
 | | X-ray crystal structure of HLA-DRA1*0101/DRB5*0101 complexed with a peptide from Epstein Barr Virus DNA polymerase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA POLYMERASE, ... | | 著者 | Lang, H, Jacobsen, H, Ikemizu, S, Andersson, C, Harlos, K, Madsen, L, Hjorth, P, Sondergaard, L, Svejgaard, A, Wucherpfennig, K, Stuart, D.I, Bell, J.I, Jones, E.Y, Fugger, L. | | 登録日 | 2002-07-02 | | 公開日 | 2002-10-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | A Functional and Structural Basis for Tcr Cross-Reactivity in Multiple Sclerosis
Nat.Immunol., 3, 2002
|
|
6NPF
 
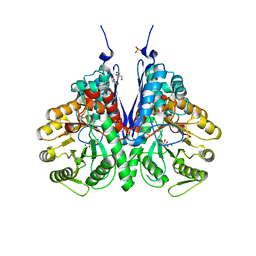 | | Structure of E.coli enolase in complex with an analog of the natural product SF-2312 metabolite. | | 分子名称: | Enolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | 著者 | Erlandsen, H, Krucinska, J, Lombardo, M, Wright, D. | | 登録日 | 2019-01-17 | | 公開日 | 2019-11-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | Functional and structural basis of E. coli enolase inhibition by SF2312: a mimic of the carbanion intermediate.
Sci Rep, 9, 2019
|
|
2NMN
 
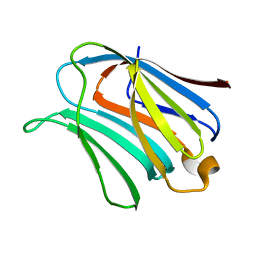 | |
7BJP
 
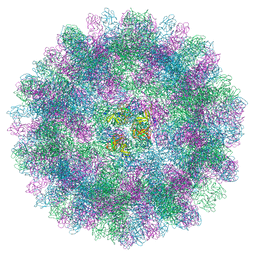 | | The cryo-EM structure of vesivirus 2117, an adventitious agent and possible cause of haemorrhagic gastroenteritis in dogs. | | 分子名称: | Capsid protein | | 著者 | Sutherland, H, Conley, M.J, Emmott, E, Streetley, J, Goodfellow, I.G, Bhella, D. | | 登録日 | 2021-01-14 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.65 Å) | | 主引用文献 | The Cryo-EM Structure of Vesivirus 2117 Highlights Functional Variations in Entry Pathways for Viruses in Different Clades of the Vesivirus Genus.
J.Virol., 95, 2021
|
|
2I2S
 
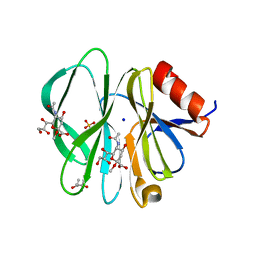 | |
2NMO
 
 | |
2NN8
 
 | | Crystal structure of human galectin-3 carbohydrate-recognition domain with lactose bound, at 1.35 angstrom resolution | | 分子名称: | CHLORIDE ION, GLYCEROL, Galectin-3, ... | | 著者 | Blanchard, H, Collins, P.M. | | 登録日 | 2006-10-24 | | 公開日 | 2007-03-06 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Slow diffusion of lactose out of galectin-3 crystals monitored by X-ray crystallography: possible implications for ligand-exchange protocols.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2P3I
 
 | | Crystal structure of Rhesus Rotavirus VP8* at 295K | | 分子名称: | 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, SULFATE ION, VP4 | | 著者 | Blanchard, H. | | 登録日 | 2007-03-09 | | 公開日 | 2008-03-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Effects on sialic acid recognition of amino acid mutations in the carbohydrate-binding cleft of the rotavirus spike protein
Glycobiology, 19, 2009
|
|
2P3K
 
 | | Crystal structure of Rhesus rotavirus VP8* at 100K | | 分子名称: | 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, GLYCEROL, SULFATE ION, ... | | 著者 | Blanchard, H. | | 登録日 | 2007-03-09 | | 公開日 | 2008-03-11 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Effects on sialic acid recognition of amino acid mutations in the carbohydrate-binding cleft of the rotavirus spike protein
Glycobiology, 19, 2009
|
|
2P3J
 
 | |
6BFZ
 
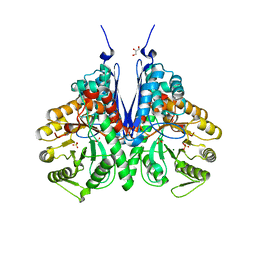 | | Crystal structure of enolase from E. coli with a mixture of apo form, substrate, and product form | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-PHOSPHOGLYCERIC ACID, Enolase, ... | | 著者 | Erlandsen, H, Wright, D, Krucinska, J. | | 登録日 | 2017-10-27 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Structural and Functional Studies of Bacterial Enolase, a Potential Target against Gram-Negative Pathogens.
Biochemistry, 58, 2019
|
|
6BFY
 
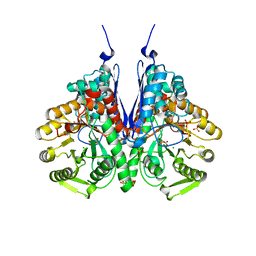 | | Crystal structure of enolase from Escherichia coli with bound 2-phosphoglycerate substrate | | 分子名称: | 2-PHOSPHOGLYCERIC ACID, Enolase, GLYCEROL, ... | | 著者 | Erlandsen, H, Wright, D, Krucinska, J. | | 登録日 | 2017-10-27 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structural and Functional Studies of Bacterial Enolase, a Potential Target against Gram-Negative Pathogens.
Biochemistry, 58, 2019
|
|
6D3Q
 
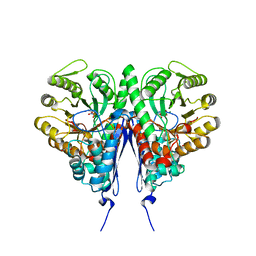 | | Crystal structure of Escherichia coli enolase complexed with a natural inhibitor SF2312. | | 分子名称: | Enolase, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Erlandsen, H, Krucinska, J, Hazeen, A, Wright, D. | | 登録日 | 2018-04-16 | | 公開日 | 2019-11-27 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Functional and structural basis of E. coli enolase inhibition by SF2312: a mimic of the carbanion intermediate.
Sci Rep, 9, 2019
|
|
6D3K
 
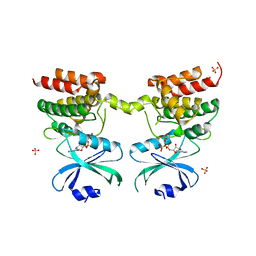 | | Crystal structure of unphosphorylated human PKR kinase domain in complex with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Interferon-induced, double-stranded RNA-activated protein kinase, ... | | 著者 | Erlandsen, H, Mayo, C.B, Robinson, V.L, Cole, J.L. | | 登録日 | 2018-04-16 | | 公開日 | 2019-07-10 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural Basis of Protein Kinase R Autophosphorylation.
Biochemistry, 58, 2019
|
|
6D3L
 
 | | Crystal structure of unphosphorylated human PKR | | 分子名称: | Interferon-induced, double-stranded RNA-activated protein kinase | | 著者 | Erlandsen, H, Mayo, C, Robinson, V.L, Cole, J.L. | | 登録日 | 2018-04-16 | | 公開日 | 2019-07-10 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural Basis of Protein Kinase R Autophosphorylation.
Biochemistry, 58, 2019
|
|
2FRX
 
 | | Crystal structure of YebU, a m5C RNA methyltransferase from E.coli | | 分子名称: | Hypothetical protein yebU | | 著者 | Erlandsen, H, Nordlund, P, Hallberg, B.M, Johnson, K.A, Ericsson, U.B. | | 登録日 | 2006-01-20 | | 公開日 | 2006-08-29 | | 最終更新日 | 2018-05-23 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The structure of the RNA m5C methyltransferase YebU from Escherichia coli reveals a C-terminal RNA-recruiting PUA domain
J.Mol.Biol., 360, 2006
|
|
4KAP
 
 | | The Binding of Benzoarylsulfonamide Ligands to Human Carbonic Anhydrase is Insensitive to Formal Fluorination of the Ligand | | 分子名称: | 4,5,6,7-tetrafluoro-1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | 著者 | Lange, H, Lockett, M.R, Breiten, B, Heroux, A, Sherman, W, Rappoport, D, Yau, P.O, Snyder, P.W, Whitesides, G.M. | | 登録日 | 2013-04-22 | | 公開日 | 2013-07-10 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | The Binding of Benzoarylsulfonamide Ligands to Human Carbonic Anhydrase is Insensitive to Formal Fluorination of the Ligand.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
2DWR
 
 | |
5PAH
 
 | |
1LTZ
 
 | | CRYSTAL STRUCTURE OF CHROMOBACTERIUM VIOLACEUM PHENYLALANINE HYDROXYLASE, STRUCTURE HAS BOUND IRON (III) AND OXIDIZED COFACTOR 7,8-DIHYDROBIOPTERIN | | 分子名称: | 7,8-DIHYDROBIOPTERIN, CHLORIDE ION, FE (III) ION, ... | | 著者 | Erlandsen, H, Kim, J.Y, Patch, M.G, Han, A, Volner, A, Abu-Omar, M.M, Stevens, R.C. | | 登録日 | 2002-05-21 | | 公開日 | 2002-07-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural comparison of bacterial and human iron-dependent phenylalanine hydroxylases: similar fold, different stability and reaction rates.
J.Mol.Biol., 320, 2002
|
|
1LTV
 
 | | CRYSTAL STRUCTURE OF CHROMOBACTERIUM VIOLACEUM PHENYLALANINE HYDROXYLASE, STRUCTURE WITH BOUND OXIDIZED Fe(III) | | 分子名称: | FE (III) ION, PHENYLALANINE-4-HYDROXYLASE | | 著者 | Erlandsen, H, Kim, J.Y, Patch, M.G, Han, A, Volner, A, Abu-Omar, M.M, Stevens, R.C. | | 登録日 | 2002-05-20 | | 公開日 | 2002-07-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural comparison of bacterial and human iron-dependent phenylalanine hydroxylases: similar fold, different stability and reaction rates.
J.Mol.Biol., 320, 2002
|
|
1LTU
 
 | | CRYSTAL STRUCTURE OF CHROMOBACTERIUM VIOLACEUM, APO (NO IRON BOUND) STRUCTURE | | 分子名称: | PHENYLALANINE-4-HYDROXYLASE | | 著者 | Erlandsen, H, Kim, J.Y, Patch, M.G, Han, A, Volner, A, Abu-Omar, M.M, Stevens, R.C. | | 登録日 | 2002-05-20 | | 公開日 | 2002-07-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Structural comparison of bacterial and human iron-dependent phenylalanine hydroxylases: similar fold, different stability and reaction rates.
J.Mol.Biol., 320, 2002
|
|
