3L3T
 
 | | Human mesotrypsin complexed with amyloid precursor protein inhibitor variant (APPIR15K) | | Descriptor: | CALCIUM ION, FORMIC ACID, PRSS3 protein, ... | | Authors: | Salameh, M.A, Soares, A.S, Radisky, E.S. | | Deposit date: | 2009-12-17 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.378 Å) | | Cite: | Determinants of affinity and proteolytic stability in interactions of Kunitz family protease inhibitors with mesotrypsin.
J.Biol.Chem., 285, 2010
|
|
3L33
 
 | | Human mesotrypsin complexed with amyloid precursor protein inhibitor(APPI) | | Descriptor: | Amyloid beta A4 protein, CALCIUM ION, FORMIC ACID, ... | | Authors: | Salameh, M.A, Soares, A.S, Radisky, E.S. | | Deposit date: | 2009-12-16 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Determinants of affinity and proteolytic stability in interactions of Kunitz family protease inhibitors with mesotrypsin.
J.Biol.Chem., 285, 2010
|
|
7JPE
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with m7GpppA Cap-0 and SAM Determined by Fixed-Target Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Non-structural protein 10, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-08-07 | | Release date: | 2020-08-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KOA
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/16 Methyltransferase in a Complex with Cap-0 and SAM Determined by Pink-Beam Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, Non-structural protein 10, P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Shuvalova, L, Lavens, A, Henning, R, Maltseva, N, Rosas-Lemus, M, Kim, Y, Satchell, K.J.F, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-07 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with Cap-0 and SAM Determined by Pink-Beam Serial Crystallography
To Be Published
|
|
7JHE
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with 2'-O-methylated m7GpppA Cap-1 and SAH Determined by Fixed-Target Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-20 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7JIB
 
 | | Room Temperature Crystal Structure of Nsp10/Nsp16 from SARS-CoV-2 with Substrates and Products of 2'-O-methylation of the Cap-1 | | Descriptor: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, ... | | Authors: | Wilamowski, M, Minasov, G, Kim, Y, Sherrell, D.A, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7L52
 
 | | Crystal Structure of the Metallo Beta Lactamase L1 from Stenotrophomonas maltophilia Determined by Serial Crystallography | | Descriptor: | Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Maltseva, N, Endres, M, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-21 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Metallo Beta Lactamase L1 from Stenotrophomonas maltophilia Determined by Serial Crystallography
To Be Published
|
|
2QI4
 
 | | Crystal structure of protease inhibitor, MIT-2-AD93 in complex with wild type HIV-1 protease | | Descriptor: | ACETATE ION, N-[(1S,2R)-3-{(1,3-BENZOTHIAZOL-6-YLSULFONYL)[(2S)-2-METHYLBUTYL]AMINO}-1-BENZYL-2-HYDROXYPROPYL]-3-HYDROXYBENZAMIDE, PHOSPHATE ION, ... | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
2QI3
 
 | | Crystal structure of protease inhibitor, MIT-2-AD94 in complex with wild type HIV-1 protease | | Descriptor: | (2S)-N-[(1S,2R)-3-{(1,3-BENZOTHIAZOL-6-YLSULFONYL)[(2S)-2-METHYLBUTYL]AMINO}-1-BENZYL-2-HYDROXYPROPYL]-2-HYDROXY-3-METHYLBUTANAMIDE, PHOSPHATE ION, Protease | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
7VOI
 
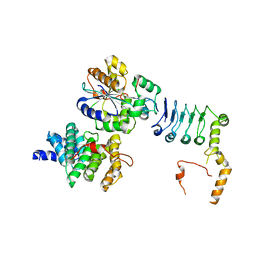 | | Structure of the human CNOT1(MIF4G)-CNOT6L-CNOT7 complex | | Descriptor: | CCR4-NOT transcription complex subunit 1, CCR4-NOT transcription complex subunit 6-like, CCR4-NOT transcription complex subunit 7 | | Authors: | Bartlam, M, Zhang, Q. | | Deposit date: | 2021-10-13 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.38 Å) | | Cite: | Structure of the human Ccr4-Not nuclease module using X-ray crystallography and electron paramagnetic resonance spectroscopy distance measurements.
Protein Sci., 31, 2022
|
|
1I2V
 
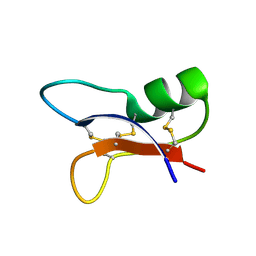 | | NMR SOLUTION STRUCTURES OF AN ANTIFUNGAL AND ANTIBACTERIAL MUTANT OF HELIOMICIN | | Descriptor: | DEFENSIN HELIOMICIN | | Authors: | Lamberty, M, Caille, A, Landon, C, Tassin-Moindrot, S, Hetru, C, Bulet, P, Vovelle, F. | | Deposit date: | 2001-02-12 | | Release date: | 2002-02-12 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the antifungal heliomicin and a selected variant with both antibacterial and antifungal activities.
Biochemistry, 40, 2001
|
|
1HAQ
 
 | |
2R9P
 
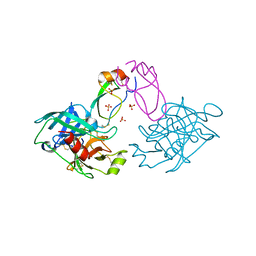 | |
8OEB
 
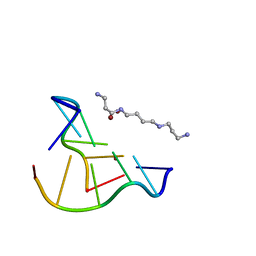 | |
8OE8
 
 | |
8OE3
 
 | |
8OE7
 
 | |
8OEX
 
 | |
8OEZ
 
 | |
8OEA
 
 | |
8OE9
 
 | |
8OEC
 
 | |
1I2U
 
 | | NMR SOLUTION STRUCTURES OF ANTIFUNGAL HELIOMICIN | | Descriptor: | DEFENSIN HELIOMICIN | | Authors: | Lamberty, M, Caille, A, Landon, C, Tassin-Moindrot, S, Hetru, C, Bulet, P, Vovelle, F. | | Deposit date: | 2001-02-12 | | Release date: | 2002-02-12 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the antifungal heliomicin and a selected variant with both antibacterial and antifungal activities.
Biochemistry, 40, 2001
|
|
1NG9
 
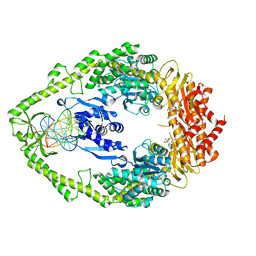 | | E.coli MutS R697A: an ATPase-asymmetry mutant | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP*GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP*GP*GP*CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lamers, M.H, Winterwerp, H.H.K, Sixma, T.K. | | Deposit date: | 2002-12-17 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The alternating ATPase domains of MutS control DNA mismatch repair
Embo J., 22, 2003
|
|
1NTL
 
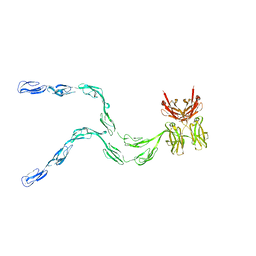 | | Model of mouse Crry-Ig determined by solution scattering, curve fitting and homology modelling | | Descriptor: | Complement component receptor 1-like protein,Ig gamma-1 chain C region secreted form | | Authors: | Aslam, M, Guthridge, J.M, Hack, B.K, Quigg, R.J, Holers, V.M, Perkins, S.J. | | Deposit date: | 2003-01-30 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | SOLUTION SCATTERING (30 Å) | | Cite: | The extended multidomain solution structures of the complement protein Crry
and its chimaeric conjugate Crry-Ig by scattering, analytical ultracentrifugation
and constrained modelling: implications for function and therapy
J.Mol.Biol., 329, 2003
|
|
