352D
 
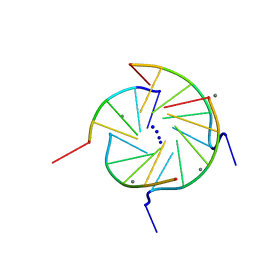 | | THE CRYSTAL STRUCTURE OF A PARALLEL-STRANDED PARALLEL-STRANDED GUANINE TETRAPLEX AT 0.95 ANGSTROM RESOLUTION | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*GP*GP*GP*GP*T)-3'), SODIUM ION | | Authors: | Phillips, K, Dauter, Z, Murchie, A.I.H, Lilley, D.M.J, Luisi, B. | | Deposit date: | 1997-09-04 | | Release date: | 1997-11-10 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | The crystal structure of a parallel-stranded guanine tetraplex at 0.95 A resolution.
J.Mol.Biol., 273, 1997
|
|
2YJT
 
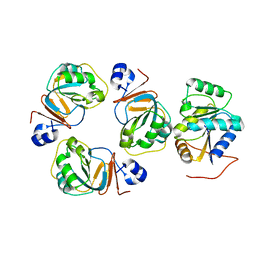 | |
4RMO
 
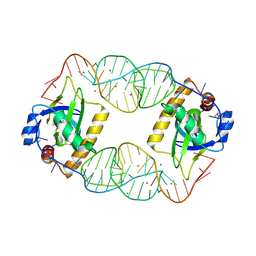 | | Crystal Structure of the CptIN Type III Toxin-Antitoxin System from Eubacterium rectale | | Descriptor: | CALCIUM ION, CptN Toxin, RNA (45-MER) | | Authors: | Rao, F, Voss, J.E, Short, F.L, Luisi, B.F. | | Deposit date: | 2014-10-21 | | Release date: | 2015-09-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Co-evolution of quaternary organization and novel RNA tertiary interactions revealed in the crystal structure of a bacterial protein-RNA toxin-antitoxin system.
Nucleic Acids Res., 43, 2015
|
|
4AID
 
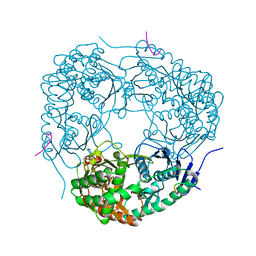 | | Crystal structure of C. crescentus PNPase bound to RNase E recognition peptide | | Descriptor: | PHOSPHATE ION, POLYRIBONUCLEOTIDE NUCLEOTIDYLTRANSFERASE, RIBONUCLEASE, ... | | Authors: | Hardwick, S.W, Gubbey, T, Hug, I, Jenal, U, Luisi, B.F. | | Deposit date: | 2012-02-09 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Caulobacter Crescentus Polynucleotide Phosphorylase Reveals a Mechanism of RNA Substrate Channelling and RNA Degradosome Assembly.
Open Biol., 2, 2012
|
|
3GME
 
 | |
7OGL
 
 | | A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation. apo-PNPase | | Descriptor: | Polyribonucleotide nucleotidyltransferase | | Authors: | Dendooven, T, Sinha, D, Roesoleva, A, Cameron, T.A, De Lay, N, Luisi, B.F, Bandyra, K. | | Deposit date: | 2021-05-06 | | Release date: | 2021-07-07 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation.
Mol.Cell, 81, 2021
|
|
7OGK
 
 | | A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation. PNPase-3'ETS(leuZ) | | Descriptor: | 3'ETS(LeuZ), Polyribonucleotide nucleotidyltransferase | | Authors: | Dendooven, T, Sinha, D, Roesoleva, A, Cameron, T.A, De Lay, N, Luisi, B.F, Bandyra, K. | | Deposit date: | 2021-05-06 | | Release date: | 2021-07-07 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation.
Mol.Cell, 81, 2021
|
|
1D98
 
 | | THE STRUCTURE OF AN OLIGO(DA).OLIGO(DT) TRACT AND ITS BIOLOGICAL IMPLICATIONS | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*AP*AP*AP*AP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*TP*TP*TP*TP*TP*GP*CP*G)-3') | | Authors: | Nelson, H.C.M, Finch, J.T, Luisi, B.F, Klug, A. | | Deposit date: | 1992-10-17 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of an oligo(dA).oligo(dT) tract and its biological implications.
Nature, 330, 1987
|
|
1TRO
 
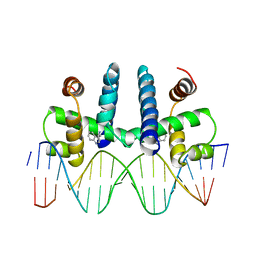 | | CRYSTAL STRUCTURE OF TRP REPRESSOR OPERATOR COMPLEX AT ATOMIC RESOLUTION | | Descriptor: | DNA (5'-D(*TP*GP*TP*AP*CP*TP*AP*GP*TP*TP*AP*AP*CP*TP*AP*GP*T P*AP*C)-3'), PROTEIN (TRP REPRESSOR), TRYPTOPHAN | | Authors: | Otwinowski, Z, Schevitz, R.W, Zhang, R.-G, Lawson, C.L, Joachimiak, A, Marmorstein, R, Luisi, B.F, Sigler, P.B. | | Deposit date: | 1992-08-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of trp repressor/operator complex at atomic resolution.
Nature, 335, 1988
|
|
1P7J
 
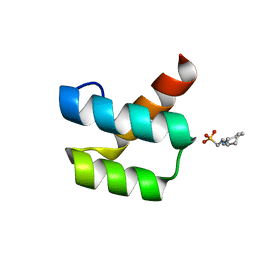 | | Crystal structure of engrailed homeodomain mutant K52E | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Segmentation polarity homeobox protein engrailed | | Authors: | Stollar, E.J, Mayor, U, Lovell, S.C, Federici, L, Freund, S.M, Fersht, A.R, Luisi, B.F. | | Deposit date: | 2003-05-02 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Engrailed Homeodomain Mutants: IMPLICATIONS FOR STABILITY AND DYNAMICS
J.Biol.Chem., 278, 2003
|
|
4AM3
 
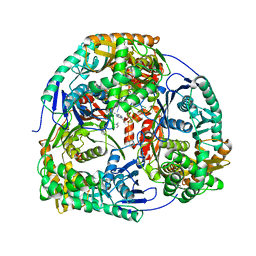 | | Crystal structure of C. crescentus PNPase bound to RNA | | Descriptor: | PHOSPHATE ION, POLYRIBONUCLEOTIDE NUCLEOTIDYLTRANSFERASE, RNA, ... | | Authors: | Hardwick, S.W, Gubbey, T, Hug, I, Jenal, U, Luisi, B.F. | | Deposit date: | 2012-03-07 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Caulobacter Crescentus Polynucleotide Phosphorylase Reveals a Mechanism of RNA Substrate Channelling and RNA Degradosome Assembly.
Open Biol., 2, 2012
|
|
1P7I
 
 | | CRYSTAL STRUCTURE OF ENGRAILED HOMEODOMAIN MUTANT K52A | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Segmentation polarity homeobox protein engrailed | | Authors: | Stollar, E.J, Mayor, U, Lovell, S.C, Federici, L, Freund, S.M, Fersht, A.R, Luisi, B.F. | | Deposit date: | 2003-05-02 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Engrailed Homeodomain Mutants: IMPLICATIONS FOR STABILITY AND DYNAMICS
J.Biol.Chem., 278, 2003
|
|
1W08
 
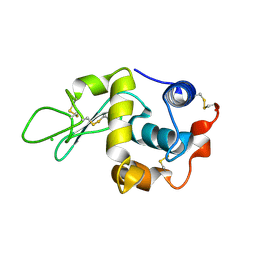 | | STRUCTURE OF T70N HUMAN LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME | | Authors: | Johnson, R, Christodoulou, J, Luisi, B, Dumoulin, M, Caddy, G, Alcocer, M, Murtagh, G, Archer, D.B, Dobson, C.M. | | Deposit date: | 2004-06-02 | | Release date: | 2004-06-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rationalising Lysozyme Amyloidosis: Insights from the Structure and Solution Dynamics of T70N Lysozyme.
J.Mol.Biol., 352, 2005
|
|
1E9I
 
 | | Enolase from E.coli | | Descriptor: | ENOLASE, MAGNESIUM ION, SULFATE ION | | Authors: | Kuhnel, K, Carpousis, A.J, Luisi, B. | | Deposit date: | 2000-10-17 | | Release date: | 2001-03-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structure of the Escherichia Coli RNA Degradosome Component Enolase
J.Mol.Biol., 313, 2001
|
|
1IBE
 
 | |
3DVA
 
 | | Snapshots of catalysis in the E1 subunit of the pyruvate dehydrogenase multi-enzyme complex | | Descriptor: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-3-METHYLTHIOPHEN-2-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, MAGNESIUM ION, ... | | Authors: | Pei, X.Y, Titman, C.M, Frank, R.A.W, Leeper, F.J, Luisi, B.F. | | Deposit date: | 2008-07-18 | | Release date: | 2009-01-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Snapshots of catalysis in the e1 subunit of the pyruvate dehydrogenase multienzyme complex
Structure, 16, 2008
|
|
3DUF
 
 | | Snapshots of catalysis in the E1 subunit of the pyruvate dehydrogenase multi-enzyme complex | | Descriptor: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-5-[(1R)-1-HYDROXYETHYL]-3-METHYL-2-THIENYL}ETHYL TRIHYDROGEN DIPHOSPHATE, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, MAGNESIUM ION, ... | | Authors: | Pei, X.Y, Titman, C.M, Frank, R.A.W, Leeper, F.J, Luisi, B.F. | | Deposit date: | 2008-07-17 | | Release date: | 2009-01-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Snapshots of catalysis in the e1 subunit of the pyruvate dehydrogenase multienzyme complex
Structure, 16, 2008
|
|
3GCM
 
 | |
3H8A
 
 | |
2YJN
 
 | | Structure of the glycosyltransferase EryCIII from the erythromycin biosynthetic pathway, in complex with its activating partner, EryCII | | Descriptor: | DTDP-4-KETO-6-DEOXY-HEXOSE 3,4-ISOMERASE, GLYCOSYLTRANSFERASE | | Authors: | Moncrieffe, M.C, Fernandez, M.J, Spiteller, D, Matsumura, H, Gay, N.J, Luisi, B.F, Leadlay, P.F. | | Deposit date: | 2011-05-20 | | Release date: | 2011-11-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.091 Å) | | Cite: | Structure of the Glycosyltransferase Eryciii in Complex with its Activating P450 Homologue Erycii.
J.Mol.Biol., 415, 2012
|
|
1YC9
 
 | | The crystal structure of the outer membrane protein VceC from the bacterial pathogen Vibrio cholerae at 1.8 resolution | | Descriptor: | MERCURY (II) ION, multidrug resistance protein, octyl beta-D-glucopyranoside | | Authors: | Federici, L, Du, D, Walas, F, Matsumura, H, Fernandez-Recio, J, McKeegan, K.S, Borges-Walmsley, M.I, Luisi, B.F, Walmsley, A.R. | | Deposit date: | 2004-12-22 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the outer membrane protein VCEC from the bacterial pathogen vibrio cholerae at 1.8 A resolution
J.Biol.Chem., 280, 2005
|
|
3DV0
 
 | | Snapshots of catalysis in the E1 subunit of the pyruvate dehydrogenase multi-enzyme complex | | Descriptor: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-3-METHYLTHIOPHEN-2-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, MAGNESIUM ION, ... | | Authors: | Pei, X.Y, Titman, C.M, Frank, R.A.W, Leeper, F.J, Luisi, B.F. | | Deposit date: | 2008-07-18 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Snapshots of catalysis in the e1 subunit of the pyruvate dehydrogenase multienzyme complex
Structure, 16, 2008
|
|
7PCH
 
 | | Human carboxyhemoglobin bound to Staphylococcus aureus hemophore IsdB - 1:2 complex | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, Iron-regulated surface determinant protein B, ... | | Authors: | De Bei, O, Gianquinto, E, Chirgadze, D.Y, Hardwick, S.W, Spyrakis, F, Luisi, B.F, Campanini, B. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM structures of staphylococcal IsdB bound to human hemoglobin reveal the process of heme extraction.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7PCF
 
 | | Human methemoglobin bound to Staphylococcus aureus hemophore IsdB | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, Iron-regulated surface determinant protein B, ... | | Authors: | De Bei, O, Gianquinto, E, Chirgadze, D.Y, Hardwick, S.W, Spyrakis, F, Luisi, B.F, Campanini, B. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (5.82 Å) | | Cite: | Cryo-EM structures of staphylococcal IsdB bound to human hemoglobin reveal the process of heme extraction.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7PCQ
 
 | | Human carboxyhemoglobin bound to Staphylococcus aureus hemophore IsdB - 1:1 complex | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, Iron-regulated surface determinant protein B, ... | | Authors: | De Bei, O, Gianquinto, E, Chirgadze, D.Y, Hardwick, S.W, Spyrakis, F, Luisi, B.F, Campanini, B. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Cryo-EM structures of staphylococcal IsdB bound to human hemoglobin reveal the process of heme extraction.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
