3GME
 
 | |
3CFS
 
 | | Structural basis of the interaction of RbAp46/RbAp48 with histone H4 | | Descriptor: | ARSENIC, Histone H4, Histone-binding protein RBBP7 | | Authors: | Murzina, N.V, Pei, X.-Y, Pratap, J.V, Sparkes, M, Vicente-Garcia, J, Ben-Shahar, T.R, Verreault, A, Luisi, B.F, Laue, E.D. | | Deposit date: | 2008-03-04 | | Release date: | 2008-06-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for the Recognition of Histone H4 by the Histone-Chaperone RbAp46.
Structure, 16, 2008
|
|
3CFV
 
 | | Structural basis of the interaction of RbAp46/RbAp48 with histone H4 | | Descriptor: | ARSENIC, Histone H4 peptide, Histone-binding protein RBBP7 | | Authors: | Pei, X.-Y, Murzina, N.V, Zhang, W, McLaughlin, S, Verreault, A, Luisi, B.F, Laue, E.D. | | Deposit date: | 2008-03-04 | | Release date: | 2008-06-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for the Recognition of Histone H4 by the Histone-Chaperone RbAp46.
Structure, 16, 2008
|
|
1TRO
 
 | | CRYSTAL STRUCTURE OF TRP REPRESSOR OPERATOR COMPLEX AT ATOMIC RESOLUTION | | Descriptor: | DNA (5'-D(*TP*GP*TP*AP*CP*TP*AP*GP*TP*TP*AP*AP*CP*TP*AP*GP*T P*AP*C)-3'), PROTEIN (TRP REPRESSOR), TRYPTOPHAN | | Authors: | Otwinowski, Z, Schevitz, R.W, Zhang, R.-G, Lawson, C.L, Joachimiak, A, Marmorstein, R, Luisi, B.F, Sigler, P.B. | | Deposit date: | 1992-08-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of trp repressor/operator complex at atomic resolution.
Nature, 335, 1988
|
|
8CBM
 
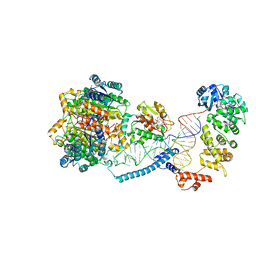 | | Structure of human mitochondrial CCA-adding enzyme in complex with mitochondrial pre-tRNA-Ile | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase type-2, CCA tRNA nucleotidyltransferase 1, mitochondrial, ... | | Authors: | MEYNIER, V, HARDWICK, S, CATALA, M, ROSKE, J, OERUM, S, CHIRGADZE, D, BARRAUD, P, LUISI, B, TISNE, C. | | Deposit date: | 2023-01-25 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis for human mitochondrial tRNA maturation.
Nat Commun, 15, 2024
|
|
8CBO
 
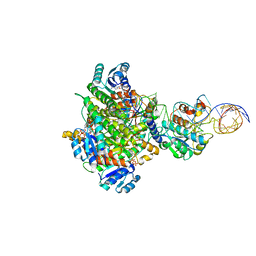 | | Structure of human mitochondrial MRPP1-MRPP2 in complex with mitochondrial pre-tRNA-Ile | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase type-2, Mitochondrial Precursor tRNA-Ile(5,4), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | MEYNIER, V, HARDWICK, S, CATALA, M, ROSKE, J, OERUM, S, CHIRGADZE, D, BARRAUD, P, LUISI, B, TISNE, C. | | Deposit date: | 2023-01-25 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for human mitochondrial tRNA maturation.
Nat Commun, 15, 2024
|
|
8CBK
 
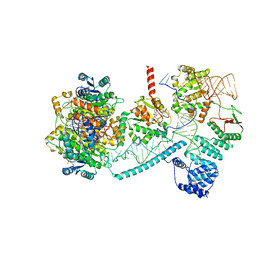 | | Structure of human mitochondrial RNase P in complex with mitochondrial pre-tRNA-His(5,Ser) | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase type-2, MAGNESIUM ION, Mitochondrial Precursor tRNA-His(5,Ser), ... | | Authors: | MEYNIER, V, HARDWICK, S, CATALA, M, ROSKE, J, OERUM, S, CHIRGADZE, D, BARRAUD, P, LUISI, B, TISNE, C. | | Deposit date: | 2023-01-25 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis for human mitochondrial tRNA maturation.
Nat Commun, 15, 2024
|
|
8CBL
 
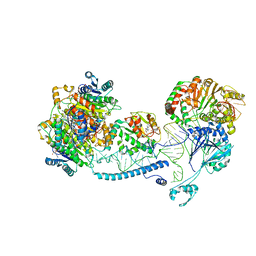 | | Structure of human mitochondrial RNase Z in complex with mitochondrial pre-tRNA-His(0,Ser) | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase type-2, Mitochondrial Precursor tRNA-His(0,Ser), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | MEYNIER, V, HARDWICK, S, CATALA, M, ROSKE, J, OERUM, S, CHIRGADZE, D, BARRAUD, P, YU, W, LUISI, B, TISNE, C. | | Deposit date: | 2023-01-25 | | Release date: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structural basis for human mitochondrial tRNA maturation.
Nat Commun, 15, 2024
|
|
4OWG
 
 | |
6TPQ
 
 | | RNase M5 bound to 50S ribosome with precursor 5S rRNA | | Descriptor: | 50S ribosomal protein L10, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Oerum, S, Dendooven, T, Gilet, L, Catala, M, Degut, C, Trinquier, A, Barraud, P, Luisi, B, Condon, C, Tisne, C. | | Deposit date: | 2019-12-13 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structures of B. subtilis Maturation RNases Captured on 50S Ribosome with Pre-rRNAs.
Mol.Cell, 80, 2020
|
|
2FIC
 
 | | The crystal structure of the BAR domain from human Bin1/Amphiphysin II and its implications for molecular recognition | | Descriptor: | Myc box-dependent-interacting protein 1, XENON | | Authors: | Casal, E, Federici, L, Zhang, W, Fernandez-Recio, J, Priego, E.M, Miguel, R.N, Duhadaway, J.B, Prendergast, G.C, Luisi, B.F, Laue, E.D. | | Deposit date: | 2005-12-29 | | Release date: | 2006-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Crystal Structure of the BAR Domain from Human Bin1/Amphiphysin II and Its Implications for Molecular Recognition
Biochemistry, 45, 2006
|
|
1W08
 
 | | STRUCTURE OF T70N HUMAN LYSOZYME | | Descriptor: | CHLORIDE ION, LYSOZYME | | Authors: | Johnson, R, Christodoulou, J, Luisi, B, Dumoulin, M, Caddy, G, Alcocer, M, Murtagh, G, Archer, D.B, Dobson, C.M. | | Deposit date: | 2004-06-02 | | Release date: | 2004-06-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rationalising Lysozyme Amyloidosis: Insights from the Structure and Solution Dynamics of T70N Lysozyme.
J.Mol.Biol., 352, 2005
|
|
3DV0
 
 | | Snapshots of catalysis in the E1 subunit of the pyruvate dehydrogenase multi-enzyme complex | | Descriptor: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-3-METHYLTHIOPHEN-2-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, MAGNESIUM ION, ... | | Authors: | Pei, X.Y, Titman, C.M, Frank, R.A.W, Leeper, F.J, Luisi, B.F. | | Deposit date: | 2008-07-18 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Snapshots of catalysis in the e1 subunit of the pyruvate dehydrogenase multienzyme complex
Structure, 16, 2008
|
|
3DVA
 
 | | Snapshots of catalysis in the E1 subunit of the pyruvate dehydrogenase multi-enzyme complex | | Descriptor: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-3-METHYLTHIOPHEN-2-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, MAGNESIUM ION, ... | | Authors: | Pei, X.Y, Titman, C.M, Frank, R.A.W, Leeper, F.J, Luisi, B.F. | | Deposit date: | 2008-07-18 | | Release date: | 2009-01-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Snapshots of catalysis in the e1 subunit of the pyruvate dehydrogenase multienzyme complex
Structure, 16, 2008
|
|
4O7J
 
 | |
3DUF
 
 | | Snapshots of catalysis in the E1 subunit of the pyruvate dehydrogenase multi-enzyme complex | | Descriptor: | 2-{4-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-5-[(1R)-1-HYDROXYETHYL]-3-METHYL-2-THIENYL}ETHYL TRIHYDROGEN DIPHOSPHATE, Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, MAGNESIUM ION, ... | | Authors: | Pei, X.Y, Titman, C.M, Frank, R.A.W, Leeper, F.J, Luisi, B.F. | | Deposit date: | 2008-07-17 | | Release date: | 2009-01-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Snapshots of catalysis in the e1 subunit of the pyruvate dehydrogenase multienzyme complex
Structure, 16, 2008
|
|
6TNN
 
 | | Mini-RNase III (Mini-III) bound to 50S ribosome with precursor 23S rRNA | | Descriptor: | 50S ribosomal protein L10, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Oerum, S, Dendooven, T, Gilet, L, Catala, M, Degut, C, Trinquier, A, Barraud, P, Luisi, B, Condon, C, Tisne, C. | | Deposit date: | 2019-12-09 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structures of B. subtilis Maturation RNases Captured on 50S Ribosome with Pre-rRNAs.
Mol.Cell, 80, 2020
|
|
1YC9
 
 | | The crystal structure of the outer membrane protein VceC from the bacterial pathogen Vibrio cholerae at 1.8 resolution | | Descriptor: | MERCURY (II) ION, multidrug resistance protein, octyl beta-D-glucopyranoside | | Authors: | Federici, L, Du, D, Walas, F, Matsumura, H, Fernandez-Recio, J, McKeegan, K.S, Borges-Walmsley, M.I, Luisi, B.F, Walmsley, A.R. | | Deposit date: | 2004-12-22 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the outer membrane protein VCEC from the bacterial pathogen vibrio cholerae at 1.8 A resolution
J.Biol.Chem., 280, 2005
|
|
2JGD
 
 | | E. COLI 2-oxoglutarate dehydrogenase (E1o) | | Descriptor: | 2-OXOGLUTARATE DEHYDROGENASE E1 COMPONENT, ADENOSINE MONOPHOSPHATE | | Authors: | Frank, R.A.W, Price, A.J, Northrop, F.D, Perham, R.N, Luisi, B.F. | | Deposit date: | 2007-02-12 | | Release date: | 2007-02-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the E1 Component of the Escherichia Coli 2-Oxoglutarate Dehydrogenase Multienzyme Complex.
J.Mol.Biol., 368, 2007
|
|
8CIB
 
 | | Structural and functional analysis of the Pseudomonas aeruginosa PA1677 protein | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Cysteine hydrolase, ... | | Authors: | Sonnleitner, E, Brear, P, Luisi, B.F, Blasi, U. | | Deposit date: | 2023-02-09 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Catabolite repression control protein antagonist, a novel player in Pseudomonas aeruginosa carbon catabolite repression control.
Front Microbiol, 14, 2023
|
|
7P06
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in outward-facing conformation with ADP-orthovanadate/ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ADP ORTHOVANADATE, MAGNESIUM ION, ... | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
7P05
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward-facing conformation with ADP/ATP and rhodamine 6G | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Pleiotropic ABC efflux transporter of multiple drugs, ... | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
7P04
 
 | | Cryo-EM structure of Pdr5 from Saccharomyces cerevisiae in inward-facing conformation with ADP/ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Pleiotropic ABC efflux transporter of multiple drugs | | Authors: | Szewczak-Harris, A, Wagner, M, Du, D, Schmitt, L, Luisi, B.F. | | Deposit date: | 2021-06-29 | | Release date: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5.
Nat Commun, 12, 2021
|
|
1P7J
 
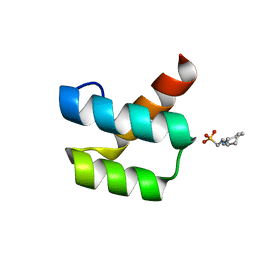 | | Crystal structure of engrailed homeodomain mutant K52E | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Segmentation polarity homeobox protein engrailed | | Authors: | Stollar, E.J, Mayor, U, Lovell, S.C, Federici, L, Freund, S.M, Fersht, A.R, Luisi, B.F. | | Deposit date: | 2003-05-02 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Engrailed Homeodomain Mutants: IMPLICATIONS FOR STABILITY AND DYNAMICS
J.Biol.Chem., 278, 2003
|
|
1P7I
 
 | | CRYSTAL STRUCTURE OF ENGRAILED HOMEODOMAIN MUTANT K52A | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Segmentation polarity homeobox protein engrailed | | Authors: | Stollar, E.J, Mayor, U, Lovell, S.C, Federici, L, Freund, S.M, Fersht, A.R, Luisi, B.F. | | Deposit date: | 2003-05-02 | | Release date: | 2003-10-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Engrailed Homeodomain Mutants: IMPLICATIONS FOR STABILITY AND DYNAMICS
J.Biol.Chem., 278, 2003
|
|
