5YJ7
 
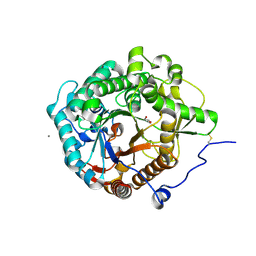 | | Structural insight into the beta-GH1 glucosidase BGLN1 from oleaginous microalgae Nannochloropsis | | 分子名称: | CALCIUM ION, GLYCEROL, Glycoside hydrolase | | 著者 | Dong, S, Liu, Y.J, Zhou, H.X, Xiao, Y, Xu, J, Cui, Q, Wang, X.Q, Feng, Y.G. | | 登録日 | 2017-10-09 | | 公開日 | 2018-10-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Structural insight into a GH1 beta-glucosidase from the oleaginous microalga, Nannochloropsis oceanica.
Int.J.Biol.Macromol., 170, 2021
|
|
8JIL
 
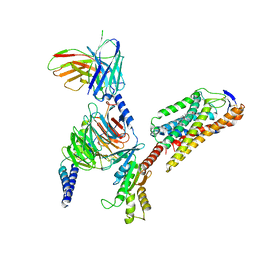 | | Cryo-EM structure of niacin bound ketone body receptor HCAR2-Gi signaling complex | | 分子名称: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Zhao, C, Tian, X.W, Liu, Y, Cheng, L, Yan, W, Shao, Z.H. | | 登録日 | 2023-05-26 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Biased allosteric activation of ketone body receptor HCAR2 suppresses inflammation.
Mol.Cell, 83, 2023
|
|
8JII
 
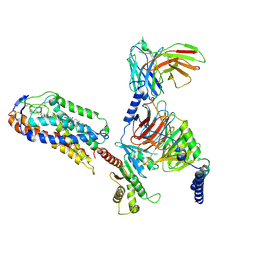 | | Cryo-EM structure of compound 9n and niacin bound ketone body receptor HCAR2-Gi signaling complex | | 分子名称: | 7-methyl-N-[(2R)-1-phenoxypropan-2-yl]-3-(4-propan-2-ylphenyl)pyrazolo[1,5-a]pyrimidine-6-carboxamide, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Zhao, C, Tian, X.W, Liu, Y, Cheng, L, Yan, W, Shao, Z.H. | | 登録日 | 2023-05-26 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | ELECTRON MICROSCOPY (3.17 Å) | | 主引用文献 | Biased allosteric activation of ketone body receptor HCAR2 suppresses inflammation.
Mol.Cell, 83, 2023
|
|
8JHY
 
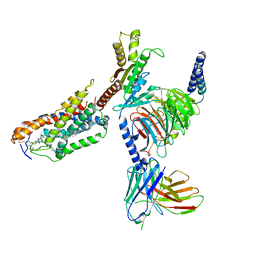 | | Cryo-EM structure of compound 9n bound ketone body receptor HCAR2-Gi signaling complex | | 分子名称: | 7-methyl-N-[(2R)-1-phenoxypropan-2-yl]-3-(4-propan-2-ylphenyl)pyrazolo[1,5-a]pyrimidine-6-carboxamide, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Zhao, C, Tian, X.W, Liu, Y, Cheng, L, Yan, W, Shao, Z.H. | | 登録日 | 2023-05-25 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | ELECTRON MICROSCOPY (2.87 Å) | | 主引用文献 | Biased allosteric activation of ketone body receptor HCAR2 suppresses inflammation.
Mol.Cell, 83, 2023
|
|
8JIM
 
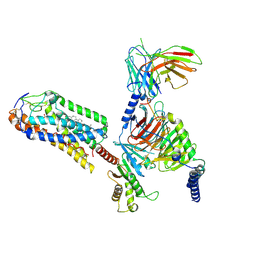 | | Cryo-EM structure of MMF bound ketone body receptor HCAR2-Gi signaling complex | | 分子名称: | (2Z)-4-methoxy-4-oxobut-2-enoic acid, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Zhao, C, Tian, X.W, Liu, Y, Cheng, L, Yan, W, Shao, Z.H. | | 登録日 | 2023-05-26 | | 公開日 | 2023-09-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | ELECTRON MICROSCOPY (2.98 Å) | | 主引用文献 | Biased allosteric activation of ketone body receptor HCAR2 suppresses inflammation.
Mol.Cell, 83, 2023
|
|
8JIF
 
 | |
7XC4
 
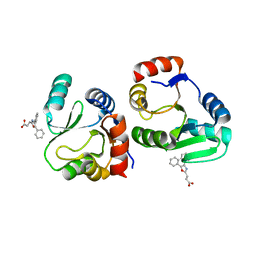 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain 3 (SARS-unique domain-M) in complex with Oxaprozin | | 分子名称: | 3-(4,5-diphenyl-1,3-oxazol-2-yl)propanoic acid, Papain-like protease nsp3 | | 著者 | Li, J, Liu, Y, Gao, J, Ruan, K. | | 登録日 | 2022-03-22 | | 公開日 | 2022-11-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Two Binding Sites of SARS-CoV-2 Macrodomain 3 Probed by Oxaprozin and Meclomen.
J.Med.Chem., 65, 2022
|
|
4PEK
 
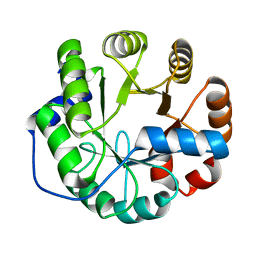 | | Crystal structure of a computationally designed retro-aldolase, RA114.3 | | 分子名称: | Retro-aldolase | | 著者 | Bhabha, G, Zhang, X, Liu, Y, Ekiert, D.C. | | 登録日 | 2014-04-23 | | 公開日 | 2015-04-08 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | De novo-designed enzymes as small-molecule-regulated fluorescence imaging tags and fluorescent reporters.
J.Am.Chem.Soc., 136, 2014
|
|
4PEJ
 
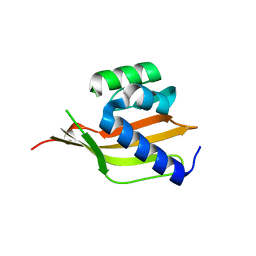 | | Crystal structure of a computationally designed retro-aldolase, RA110.4 (Cys free) | | 分子名称: | Retro-aldolase | | 著者 | Bhabha, G, Zhang, X, Liu, Y, Ekiert, D.C. | | 登録日 | 2014-04-23 | | 公開日 | 2015-04-08 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | De novo-designed enzymes as small-molecule-regulated fluorescence imaging tags and fluorescent reporters.
J.Am.Chem.Soc., 136, 2014
|
|
4EQK
 
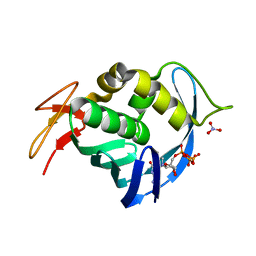 | |
7W7P
 
 | |
7X8M
 
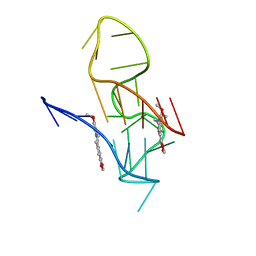 | | NMR Solution Structure of the 2:1 Berberine-KRAS-G4 Complex | | 分子名称: | BERBERINE, DNA (24-MER) | | 著者 | Wang, K.B, Liu, Y, Li, J, Xiao, C, Gu, W, Li, Y, Xia, Y.Z, Yan, T, Yang, M.H, Kong, L.Y. | | 登録日 | 2022-03-14 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural insight into the bulge-containing KRAS oncogene promoter G-quadruplex bound to berberine and coptisine.
Nat Commun, 13, 2022
|
|
5ZBM
 
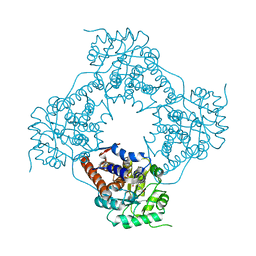 | |
4NRM
 
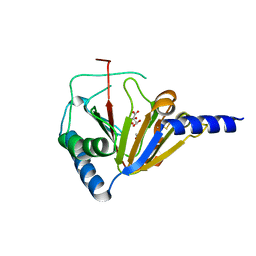 | |
6JWE
 
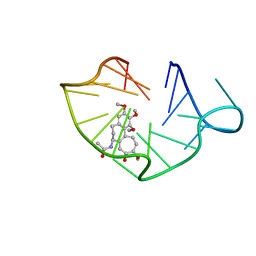 | | structure of RET G-quadruplex in complex with colchicine | | 分子名称: | DNA (5'-D(*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3'), N-[(7S)-1,2,3,10-tetramethoxy-9-oxo-6,7-dihydro-5H-benzo[d]heptalen-7-yl]ethanamide | | 著者 | Wang, F, Wang, C, Liu, Y, Lan, W.X, Li, Y.M, Wang, R.X, Cao, C. | | 登録日 | 2019-04-20 | | 公開日 | 2020-02-26 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Colchicine selective interaction with oncogene RET G-quadruplex revealed by NMR.
Chem.Commun.(Camb.), 56, 2020
|
|
6JWD
 
 | | structure of RET G-quadruplex in complex with berberine | | 分子名称: | BERBERINE, DNA (5'-D(*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3') | | 著者 | Wang, F, Wang, C, Liu, Y, Lan, W.X, Li, Y.M, Wang, R.X, Cao, C. | | 登録日 | 2019-04-19 | | 公開日 | 2020-04-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Colchicine selective interaction with oncogene RET G-quadruplex revealed by NMR.
Chem.Commun.(Camb.), 56, 2020
|
|
4GIP
 
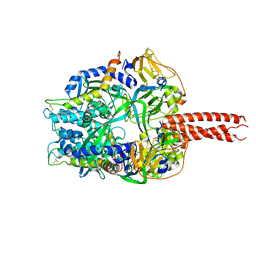 | | Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 (PIV5) fusion protein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F1, Fusion glycoprotein F2 | | 著者 | Welch, B.D, Liu, Y, Kors, C.A, Leser, G.P, Jardetzky, T.S, Lamb, R.A. | | 登録日 | 2012-08-08 | | 公開日 | 2012-09-19 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 fusion protein.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2IS9
 
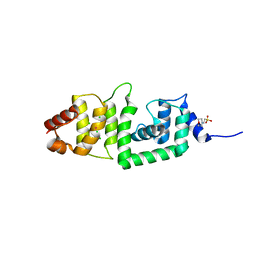 | | Structure of yeast DCN-1 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Defective in cullin neddylation protein 1, ... | | 著者 | Yang, X, Zhou, J, Sun, L, Wei, Z, Gao, J, Gong, W, Xu, R.M, Rao, Z, Liu, Y. | | 登録日 | 2006-10-16 | | 公開日 | 2007-06-26 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structural basis for the function of DCN-1 in protein Neddylation.
J.Biol.Chem., 282, 2007
|
|
4NRP
 
 | |
4ES5
 
 | |
4H5Y
 
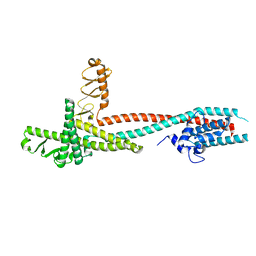 | | High-resolution crystal structure of Legionella pneumophila LidA (60-594) | | 分子名称: | LidA protein, substrate of the Dot/Icm system | | 著者 | An, X, Ye, S, Liu, Y, Zheng, X, Zhang, R. | | 登録日 | 2012-09-19 | | 公開日 | 2013-09-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The crystal structure of LidA, a translocated substrate of the Legionella pneumophila type IV secretion system.
Protein Cell, 4, 2013
|
|
4HA7
 
 | |
4HA9
 
 | |
4NRO
 
 | |
4NRQ
 
 | | Crystal structure of human ALKBH5 in complex with pyridine-2,4-dicarboxylate | | 分子名称: | MANGANESE (II) ION, PYRIDINE-2,4-DICARBOXYLIC ACID, RNA demethylase ALKBH5 | | 著者 | Feng, C, Chen, Z, Liu, Y. | | 登録日 | 2013-11-27 | | 公開日 | 2014-03-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structures of the human RNA demethylase Alkbh5 reveal basis for substrate recognition
J.Biol.Chem., 289, 2014
|
|
