7KBG
 
 | |
1ZJ6
 
 | | Crystal structure of human ARL5 | | Descriptor: | ADP-ribosylation factor-like protein 5, GUANOSINE-3'-MONOPHOSPHATE-5'-DIPHOSPHATE, SULFATE ION | | Authors: | Wang, Z.X, Shi, L, Liu, J.F, An, X.M, Chang, W.R, Liang, D.C. | | Deposit date: | 2005-04-28 | | Release date: | 2005-06-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.0A crystal structure of human ARL5-GDP3'P, a novel member of the small GTP-binding proteins
Biochem.Biophys.Res.Commun., 332, 2005
|
|
5CH8
 
 | | Crystal structure of MDLA N225Q mutant form Penicillium cyclopium | | Descriptor: | CHLORIDE ION, GLYCEROL, Mono- and diacylglycerol lipase, ... | | Authors: | Xu, J, Xu, H, Hou, S, Liu, J. | | Deposit date: | 2015-07-10 | | Release date: | 2016-04-20 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Lipase-Driven Epoxidation Is A Two-Stage Synergistic Process
ChemistrySelect, 4, 2016
|
|
8J5D
 
 | | Cryo-EM structure of starch degradation complex of BAM1-LSF1-MDH | | Descriptor: | Beta-amylase 1, chloroplastic, Malate dehydrogenase, ... | | Authors: | Guan, Z.Y, Liu, J, Yan, J.J. | | Deposit date: | 2023-04-21 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The LIKE SEX FOUR 1-malate dehydrogenase complex functions as a scaffold to recruit beta-amylase to promote starch degradation.
Plant Cell, 36, 2023
|
|
2ICT
 
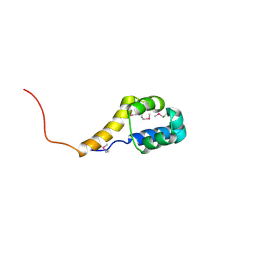 | | Crystal structure of the bacterial antitoxin HigA from Escherichia coli at pH 8.5. Northeast Structural Genomics TARGET ER390. | | Descriptor: | antitoxin higa | | Authors: | Arbing, M.A, Abashidze, M, Hurley, J.M, Zhao, L, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Inouye, M, Woychik, N.A, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-09-26 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structures of Phd-Doc, HigA, and YeeU Establish Multiple Evolutionary Links between Microbial Growth-Regulating Toxin-Antitoxin Systems.
Structure, 18, 2010
|
|
2INW
 
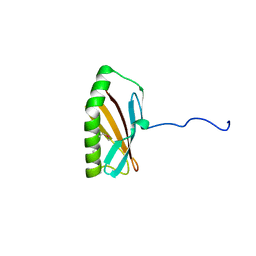 | | Crystal structure of Q83JN9 from Shigella flexneri at high resolution. Northeast Structural Genomics Consortium target SfR137. | | Descriptor: | PHOSPHATE ION, Putative structural protein | | Authors: | Kuzin, A.P, Su, M, Jayaraman, S, Vorobiev, S.M, Wang, D, Jiang, M, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-10-09 | | Release date: | 2006-10-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of Phd-Doc, HigA, and YeeU Establish Multiple Evolutionary Links between Microbial Growth-Regulating Toxin-Antitoxin Systems.
Structure, 18, 2010
|
|
3AOF
 
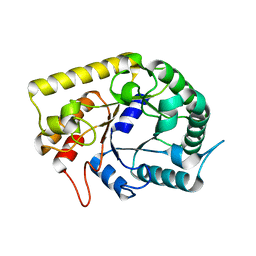 | | Crystal structures of Thermotoga maritima Cel5A in complex with Mannotriose substrate | | Descriptor: | Endoglucanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-09-27 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.288 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
7DV8
 
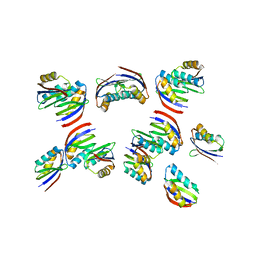 | | The crystal structure of rice immune receptor RGA5-HMA2. | | Descriptor: | Disease resistance protein RGA5 | | Authors: | Zhang, X, Liu, J.F. | | Deposit date: | 2021-01-12 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.447 Å) | | Cite: | A designer rice NLR immune receptor confers resistance to the rice blast fungus carrying noncorresponding avirulence effectors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3AMC
 
 | | Crystal structures of Thermotoga maritima Cel5A, apo form and dimer/au | | Descriptor: | Endoglucanase | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-08-19 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AMG
 
 | | Crystal structures of Thermotoga maritima Cel5A in complex with Cellobiose substrate, mutant form | | Descriptor: | Endoglucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-08-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AMD
 
 | | Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au | | Descriptor: | Endoglucanase | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-08-19 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AZR
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellobiose | | Descriptor: | Endoglucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AZS
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Mannotriose | | Descriptor: | Endoglucanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3F0S
 
 | | Staphylococcus aureus dihydrofolate reductase complexed with NADPH and 2,4-Diamino-5-[3-(3-methoxy-5-(3,5-dimethylphenyl)phenyl)but-1-ynyl]-6-methylpyrimidine | | Descriptor: | 5-[(3R)-3-(5-methoxy-3',5'-dimethylbiphenyl-3-yl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Trimethoprim-sensitive dihydrofolate reductase | | Authors: | Anderson, A.C, Frey, K.M, Liu, J, Lombardo, M.N. | | Deposit date: | 2008-10-25 | | Release date: | 2009-10-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic Complexes of Wildtype and Mutant MRSA DHFR Reveal Interactions for Lead Design
To be Published
|
|
4OD0
 
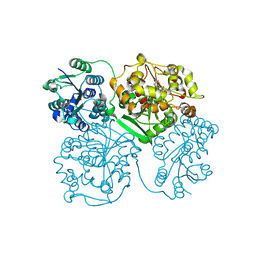 | | Crystal structure of human soluble epoxide hydrolase complexed with 1-(1-propanoylpiperidin-4-yl)-3-[4-(trifluoromethoxy)phenyl]urea | | Descriptor: | 1-(1-propanoylpiperidin-4-yl)-3-[4-(trifluoromethoxy)phenyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Lee, K.S.S, Liu, J, Wagner, K.M, Pakhomova, S, Dong, H, Morisseau, C, Fu, S.H, Yang, J, Wang, P, Ulu, A, Mate, C, Nguyen, L, Wullf, H, Eldin, M.L, Mara, A.A, Newcomer, M.E, Zeldin, D.C, Hammock, B.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Optimized inhibitors of soluble epoxide hydrolase improve in vitro target residence time and in vivo efficacy.
J.Med.Chem., 57, 2014
|
|
5VZL
 
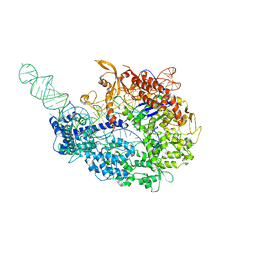 | | cryo-EM structure of the Cas9-sgRNA-AcrIIA4 anti-CRISPR complex | | Descriptor: | CRISPR-associated endonuclease Cas9, phage anti-CRISPR AcrIIA4, single guide RNA (116-MER) | | Authors: | Jiang, F, Liu, J.J, Nogales, E, Doudna, J.A. | | Deposit date: | 2017-05-29 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Disabling Cas9 by an anti-CRISPR DNA mimic.
Sci Adv, 3, 2017
|
|
5TTG
 
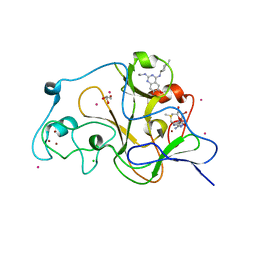 | | Crystal structure of catalytic domain of GLP with MS012 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | DONG, A, ZENG, H, LIU, J, XIONG, Y, BABAULT, N, JIN, J, TEMPEL, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, WU, H, BROWN, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-11-03 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery of Potent and Selective Inhibitors for G9a-Like Protein (GLP) Lysine Methyltransferase.
J. Med. Chem., 60, 2017
|
|
5V9I
 
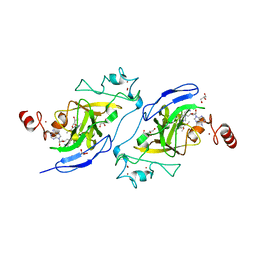 | | Crystal structure of catalytic domain of G9a with MS0105 | | Descriptor: | GLYCEROL, Histone-lysine N-methyltransferase EHMT2, N~2~-cyclohexyl-N~4~-(1-ethylpiperidin-4-yl)-6,7-dimethoxy-N~2~-methylquinazoline-2,4-diamine, ... | | Authors: | Dong, A, Zeng, H, Liu, J, Xiong, Y, Babault, N, Jin, J, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Wu, H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-23 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of catalytic domain of G9a with MS0105
to be published
|
|
4FAO
 
 | | Specificity and Structure of a high affinity Activin-like 1 (ALK1) signaling complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-2B, Growth/differentiation factor 2, ... | | Authors: | Townson, S.A, Martinez-Hackert, E, Greppi, C, Lowden, P, Sako, D, Liu, J, Ucran, J.A, Liharska, K, Underwood, K.W, Seehra, J, Kumar, R, Grinberg, A.V. | | Deposit date: | 2012-05-22 | | Release date: | 2012-06-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.357 Å) | | Cite: | Specificity and Structure of a High Affinity Activin Receptor-like Kinase 1 (ALK1) Signaling Complex.
J.Biol.Chem., 287, 2012
|
|
4OCZ
 
 | | Crystal structure of human soluble epoxide hydrolase complexed with 1-(1-isobutyrylpiperidin-4-yl)-3-(4-(trifluoromethyl)phenyl)urea | | Descriptor: | 1-[1-(2-methylpropanoyl)piperidin-4-yl]-3-[4-(trifluoromethyl)phenyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Lee, K.S.S, Liu, J, Wagner, K.M, Pakhomova, S, Dong, H, Morriseau, C, Fu, S.H, Yang, J, Wang, P, Ulu, A, Mate, C, Nguyen, L, Wullf, H, Eldin, M.L, Mara, A.A, Newcomer, M.E, Zeldin, D.C, Hammock, B.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Optimized inhibitors of soluble epoxide hydrolase improve in vitro target residence time and in vivo efficacy.
J.Med.Chem., 57, 2014
|
|
1YJ3
 
 | | Crystal structure analysis of product bound methionine aminopeptidase Type 1c from Mycobacterium Tuberculosis | | Descriptor: | BETA-MERCAPTOETHANOL, COBALT (II) ION, METHIONINE, ... | | Authors: | Addlagatta, A, Quillin, M.L, Omotoso, O, Liu, J.O, Matthews, B.W. | | Deposit date: | 2005-01-13 | | Release date: | 2005-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of an SH3-binding motif in a new class of methionine aminopeptidases from Mycobacterium tuberculosis suggests a mode of interaction with the ribosome.
Biochemistry, 44, 2005
|
|
3F0X
 
 | | Staphylococcus aureus F98Y mutant dihydrofolate reductase complexed with NADPH and 2,4-Diamino-5-[3-(3-methoxy-5-(3,5-dimethylphenyl)phenyl)but-1-ynyl]-6-methylpyrimidine | | Descriptor: | 5-[(3R)-3-(5-methoxy-3',5'-dimethylbiphenyl-3-yl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Trimethoprim-sensitive dihydrofolate reductase | | Authors: | Anderson, A.C, Frey, K.M, Liu, J, Lombardo, M.N. | | Deposit date: | 2008-10-27 | | Release date: | 2009-10-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic Complexes of Wildtype and Mutant MRSA DHFR Reveal Interactions for Lead Design
To be Published
|
|
3BD9
 
 | | human 3-O-sulfotransferase isoform 5 with bound PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Heparan sulfate glucosamine 3-O-sulfotransferase 5 | | Authors: | Xu, D, Moon, A.F, Song, D, Liu, J, Pedersen, L.C. | | Deposit date: | 2007-11-14 | | Release date: | 2008-01-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Engineering sulfotransferases to modify heparan sulfate.
Nat.Chem.Biol., 4, 2008
|
|
3F0V
 
 | | Staphylococcus aureus F98Y mutant dihydrofolate reductase complexed with NADPH and 2,4-Diamino-5-[3-(3-methoxy-5-(2,6-dimethylphenyl)phenyl)but-1-ynyl]-6-methylpyrimidine | | Descriptor: | 5-[(3S)-3-(5-methoxy-2',6'-dimethylbiphenyl-3-yl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Trimethoprim-sensitive dihydrofolate reductase | | Authors: | Anderson, A.C, Frey, K.M, Liu, J, Lombardo, M.N. | | Deposit date: | 2008-10-26 | | Release date: | 2009-10-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystallographic Complexes of Wildtype and Mutant MRSA DHFR Reveal Interactions for Lead Design
To be Published
|
|
1Y1N
 
 | | Identification of SH3 motif in M. Tuberculosis methionine aminopeptidase suggests a mode of interaction with the ribosome | | Descriptor: | Methionine aminopeptidase 1B, POTASSIUM ION | | Authors: | Addlagatta, A, Quillin, M.L, Omotoso, O, Liu, J.O, Matthews, B.W. | | Deposit date: | 2004-11-18 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Identification of an SH3-Binding Motif in a New Class of Methionine Aminopeptidases from Mycobacterium tuberculosis Suggests a Mode of Interaction with the Ribosome
Biochemistry, 44, 2005
|
|
