4R9V
 
 | | Crystal structure of sialyltransferase from photobacterium damselae, residues 113-497 corresponding to the gt-b domain | | Descriptor: | CALCIUM ION, Sialyltransferase 0160 | | Authors: | Li, Y, Huynh, N, Chen, X, Fisher, A.J. | | Deposit date: | 2014-09-08 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of sialyltransferase from Photobacterium damselae.
Febs Lett., 588, 2014
|
|
4YZE
 
 | |
6A6B
 
 | | cryo-em structure of alpha-synuclein fiber | | Descriptor: | Alpha-synuclein | | Authors: | Li, Y.W, Zhao, C.Y, Luo, F, Liu, Z, Gui, X, Luo, Z, Zhang, X, Li, D, Liu, C, Li, X. | | Deposit date: | 2018-06-27 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Amyloid fibril structure of alpha-synuclein determined by cryo-electron microscopy
Cell Res., 28, 2018
|
|
7BRE
 
 | | The crystal structure of MLL2 in complex with ASH2L and RBBP5 | | Descriptor: | Histone-lysine N-methyltransferase 2B, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Li, Y, Zhao, L, Chen, Y. | | Deposit date: | 2020-03-28 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Crystal Structure of MLL2 Complex Guides the Identification of a Methylation Site on P53 Catalyzed by KMT2 Family Methyltransferases.
Structure, 28, 2020
|
|
1NKN
 
 | | VISUALIZING AN UNSTABLE COILED COIL: THE CRYSTAL STRUCTURE OF AN N-TERMINAL SEGMENT OF THE SCALLOP MYOSIN ROD | | Descriptor: | S2N51-GCN4 | | Authors: | Li, Y, Brown, J.H, Reshetnikova, L, Blazsek, A, Farkas, L, Nyitray, L, Cohen, C. | | Deposit date: | 2003-01-03 | | Release date: | 2003-07-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Visualization of an unstable coiled coil from the scallop myosin rod
Nature, 424, 2003
|
|
8XEW
 
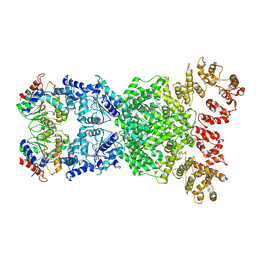 | | Cryo-EM structure of defence-associatedsirtuin 2 (DSR2) H171A protein | | Descriptor: | DSR2 H171A | | Authors: | Li, Y, Zhang, H, Zheng, Q, Wu, Y, Li, S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structural insights into activation mechanisms on NADase of the bacterial DSR2 anti-phage defense system.
Sci Adv, 10, 2024
|
|
8XFF
 
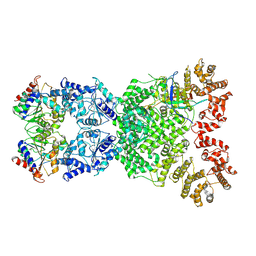 | | Cryo-EM structure of defence-associatedsirtuin 2 (DSR2) H171A protein in complex with SPR phage tail tube protein | | Descriptor: | DSR2(H171A), SPR tail tube protein | | Authors: | Li, Y, Zhang, H, Zheng, Q, Wu, Y, Li, S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural insights into activation mechanisms on NADase of the bacterial DSR2 anti-phage defense system.
Sci Adv, 10, 2024
|
|
8XFE
 
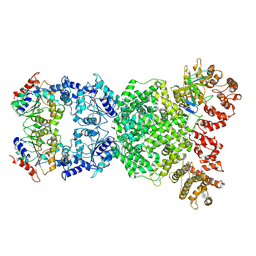 | | Cryo-EM structure of defence-associated sirtuin 2 (DSR2) H171A protein in complex with DSR anti-defence 1(DSAD1) | | Descriptor: | DSAD1, DSR2(H171A) | | Authors: | Li, Y, Zhang, H, Zheng, Q, Wu, Y, Li, S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural insights into activation mechanisms on NADase of the bacterial DSR2 anti-phage defense system.
Sci Adv, 10, 2024
|
|
4ZVV
 
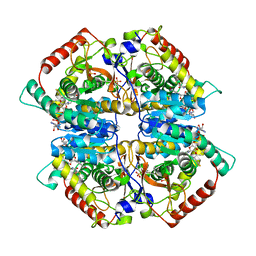 | | Lactate dehydrogenase A in complex with a trisubstituted piperidine-2,4-dione inhibitor GNE-140 | | Descriptor: | (2~{R})-5-(2-chlorophenyl)sulfanyl-2-(4-morpholin-4-ylphenyl)-4-oxidanyl-2-thiophen-3-yl-1,3-dihydropyridin-6-one, L-lactate dehydrogenase A chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Li, Y, Chen, Z, Eigenbrot, C. | | Deposit date: | 2015-05-18 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Metabolic plasticity underpins innate and acquired resistance to LDHA inhibition.
Nat.Chem.Biol., 12, 2016
|
|
5F59
 
 | | The crystal structure of MLL3 SET domain | | Descriptor: | Histone-lysine N-methyltransferase 2C, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Li, Y, Lei, M, Chen, Y. | | Deposit date: | 2015-12-04 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural basis for activity regulation of MLL family methyltransferases.
Nature, 530, 2016
|
|
1XOP
 
 | | NMR structure of G1V mutant of influenza hemagglutinin fusion peptide in DPC micelles at pH 5 | | Descriptor: | Hemagglutinin | | Authors: | Li, Y, Han, X, Lai, A.L, Bushweller, J.H, Cafiso, D.S, Tamm, L.K. | | Deposit date: | 2004-10-06 | | Release date: | 2005-09-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Membrane structures of the hemifusion-inducing fusion peptide mutant G1S and the fusion-blocking mutant G1V of influenza virus hemagglutinin suggest a mechanism for pore opening in membrane fusion.
J.Virol., 79, 2005
|
|
5F6L
 
 | | The crystal structure of MLL1 (N3861I/Q3867L) in complex with RbBP5 and Ash2L | | Descriptor: | Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Li, Y, Lei, M, Chen, Y. | | Deposit date: | 2015-12-06 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for activity regulation of MLL family methyltransferases.
Nature, 530, 2016
|
|
5F5E
 
 | |
1XOO
 
 | | NMR structure of G1S mutant of influenza hemagglutinin fusion peptide in DPC micelles at pH 5 | | Descriptor: | Hemagglutinin | | Authors: | Li, Y, Han, X, Lai, A.L, Bushweller, J.H, Cafiso, D.S, Tamm, L.K. | | Deposit date: | 2004-10-06 | | Release date: | 2005-09-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Membrane structures of the hemifusion-inducing fusion peptide mutant G1S and the fusion-blocking mutant G1V of influenza virus hemagglutinin suggest a mechanism for pore opening in membrane fusion.
J.Virol., 79, 2005
|
|
5F6K
 
 | | Crystal structure of the MLL3-Ash2L-RbBP5 complex | | Descriptor: | Histone-lysine N-methyltransferase 2C, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Li, Y, Lei, M, Chen, Y. | | Deposit date: | 2015-12-06 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Structural basis for activity regulation of MLL family methyltransferases.
Nature, 530, 2016
|
|
1XAW
 
 | |
1YP0
 
 | | Structure of the steroidogenic factor-1 ligand binding domain bound to phospholipid and a SHP peptide motif | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Nuclear receptor subfamily 0, group B, ... | | Authors: | Li, Y, Choi, M, Cavey, G, Daugherty, J, Suino, K, Kovach, A, Bingham, N, Kliewer, S, Xu, H. | | Deposit date: | 2005-01-28 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic identification and functional characterization of phospholipids as ligands for the orphan nuclear receptor steroidogenic factor-1.
Mol.Cell, 17, 2005
|
|
8JA5
 
 | | Crystal structure of Nipah Virus attachment (G) glycoprotein in complex with neutralizing antibody 14F8 | | Descriptor: | 14F8 antibody heavy chain, 14F8 antibody light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, Y.H, Huang, X.Y, Xu, J.J, Chen, W. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of Nipah Virus attachment (G) glycoprotein in complex with neutralizing antibody 14F8
To Be Published
|
|
7YTC
 
 | | Cryo-EM structure of human FcmR bound to IgM-Fc/J | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fas apoptotic inhibitory molecule 3, Immunoglobulin J chain, ... | | Authors: | Li, Y, Shen, H, Xiao, J. | | Deposit date: | 2022-08-14 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Immunoglobulin M perception by Fc mu R.
Nature, 615, 2023
|
|
1QSS
 
 | | DDGTP-TRAPPED CLOSED TERNARY COMPLEX OF THE LARGE FRAGMENT OF DNA POLYMERASE I FROM THERMUS AQUATICUS | | Descriptor: | 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*AP*CP*CP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3', 5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DDG))-3', ... | | Authors: | Li, Y, Mitaxov, V, Waksman, G. | | Deposit date: | 1999-06-23 | | Release date: | 1999-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based design of Taq DNA polymerases with improved properties of dideoxynucleotide incorporation.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3BW3
 
 | | Crystal structures and site-directed mutagenesis study of nitroalkane oxidase from Streptomyces ansochromogenes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-nitropropane dioxygenase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Li, Y.H, Gao, Z.Q, Hou, H.F. | | Deposit date: | 2008-01-08 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and site-directed mutagenesis of a nitroalkane oxidase from Streptomyces ansochromogenes
Biochem.Biophys.Res.Commun., 405, 2011
|
|
8GPT
 
 | | YFV_E_YD6scFv_postfusion | | Descriptor: | Envelope protein, YD6_VH, YD6_VL | | Authors: | Li, Y, Wu, L, Qi, J, Yan, J, Gao, G.F. | | Deposit date: | 2022-08-27 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | A neutralizing-protective supersite of human monoclonal antibodies for yellow fever virus.
Innovation (N Y), 3, 2022
|
|
3BW2
 
 | | Crystal structures and site-directed mutagenesis study of nitroalkane oxidase from Streptomyces ansochromogenes | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-nitropropane dioxygenase, FLAVIN MONONUCLEOTIDE | | Authors: | Li, Y.H, Gao, Z.Q, Hou, H.F. | | Deposit date: | 2008-01-08 | | Release date: | 2009-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and site-directed mutagenesis of a nitroalkane oxidase from Streptomyces ansochromogenes
Biochem.Biophys.Res.Commun., 405, 2011
|
|
8W7J
 
 | | Cryo-EM structure of ClassIII Lanthipeptide modification enzyme PneKC with chain A bounded to substrate PneA and GTP. | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, PneA LP, ... | | Authors: | Li, Y, Luo, M, Shao, K, Li, J, Li, Z. | | Deposit date: | 2023-08-30 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Mechanistic insights into lanthipeptide modification by a distinct subclass of LanKC enzyme that forms dimers.
Nat Commun, 15, 2024
|
|
4N4H
 
 | | Crystal structure of the Bromo-PWWP of the mouse zinc finger MYND-type containing 11 isoform alpha in complex with histone H3.1K36me3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Peptide from Histone H3.1, ... | | Authors: | Li, Y, Ren, Y, Li, H. | | Deposit date: | 2013-10-08 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression
Nature, 508, 2014
|
|
