7EP2
 
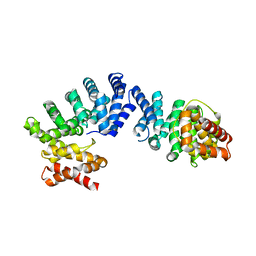 | | Crystal structure of ZYG11B bound to GGFN degron | | Descriptor: | Protein zyg-11 homolog B | | Authors: | Yan, X, Li, Y. | | Deposit date: | 2021-04-26 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Molecular basis for recognition of Gly/N-degrons by CRL2 ZYG11B and CRL2 ZER1 .
Mol.Cell, 81, 2021
|
|
7EP5
 
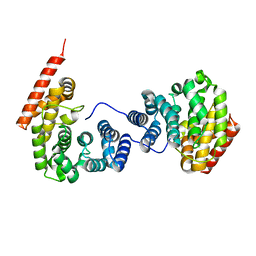 | | Crystal structure of ZER1 bound to GKLH degron | | Descriptor: | Protein zer-1 homolog | | Authors: | Yan, X, Li, Y. | | Deposit date: | 2021-04-26 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Molecular basis for recognition of Gly/N-degrons by CRL2 ZYG11B and CRL2 ZER1 .
Mol.Cell, 81, 2021
|
|
7EP1
 
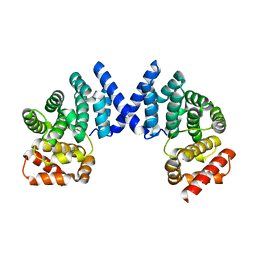 | | Crystal structure of ZYG11B bound to GFLH degron | | Descriptor: | Protein zyg-11 homolog B | | Authors: | Yan, X, Li, Y. | | Deposit date: | 2021-04-26 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Molecular basis for recognition of Gly/N-degrons by CRL2 ZYG11B and CRL2 ZER1 .
Mol.Cell, 81, 2021
|
|
7EP4
 
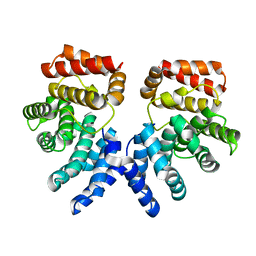 | | Crystal structure of ZER1 bound to GFLH degron | | Descriptor: | Protein zer-1 homolog | | Authors: | Yan, X, Li, Y. | | Deposit date: | 2021-04-26 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Molecular basis for recognition of Gly/N-degrons by CRL2 ZYG11B and CRL2 ZER1 .
Mol.Cell, 81, 2021
|
|
2J0J
 
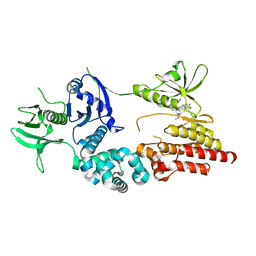 | | Crystal structure of a fragment of focal adhesion kinase containing the FERM and kinase domains. | | Descriptor: | 1,2,3,4-TETRAHYDROGEN-STAUROSPORINE, FOCAL ADHESION KINASE 1 | | Authors: | Lietha, D, Cai, X, Li, Y, Schaller, M.D, Eck, M.J. | | Deposit date: | 2006-08-03 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Autoinhibition of Focal Adhesion Kinase
Cell(Cambridge,Mass.), 129, 2007
|
|
2J0L
 
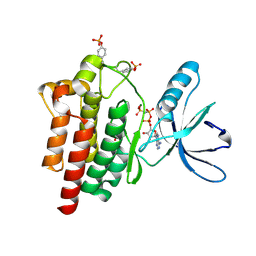 | | Crystal structure of a the active conformation of the kinase domain of focal adhesion kinase with a phosphorylated activation loop. | | Descriptor: | FOCAL ADHESION KINASE 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Lietha, D, Cai, X, Li, Y, Schaller, M.D, Eck, M.J. | | Deposit date: | 2006-08-03 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for the Autoinhibition of Focal Adhesion Kinase
Cell(Cambridge,Mass.), 129, 2007
|
|
2ITQ
 
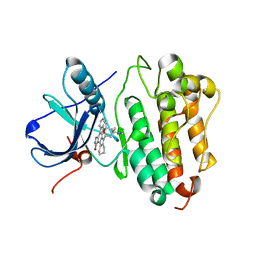 | | Crystal structure of EGFR kinase domain G719S mutation in complex with AFN941 | | Descriptor: | 1,2,3,4-Tetrahydrogen Staurosporine, EPIDERMAL GROWTH FACTOR RECEPTOR | | Authors: | Yun, C.-H, Boggon, T.J, Li, Y, Woo, S, Greulich, H, Meyerson, M, Eck, M.J. | | Deposit date: | 2006-05-25 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structures of Lung Cancer-Derived Egfr Mutants and Inhibitor Complexes: Mechanism of Activation and Insights Into Differential Inhibitor Sensitivity
Cancer Cell, 11, 2007
|
|
2ITV
 
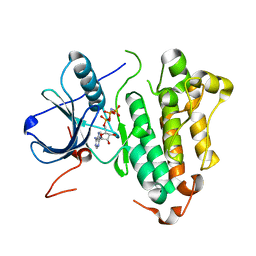 | | Crystal structure of EGFR kinase domain L858R mutation in complex with AMP-PNP | | Descriptor: | EPIDERMAL GROWTH FACTOR RECEPTOR, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yun, C.-H, Boggon, T.J, Li, Y, Woo, S, Greulich, H, Meyerson, M, Eck, M.J. | | Deposit date: | 2006-05-25 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structures of Lung Cancer-Derived Egfr Mutants and Inhibitor Complexes: Mechanism of Activation and Insights Into Differential Inhibitor Sensitivity
Cancer Cell, 11, 2007
|
|
2J6M
 
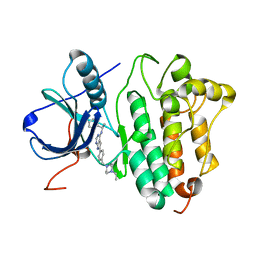 | | Crystal structure of EGFR kinase domain in complex with AEE788 | | Descriptor: | 6-{4-[(4-ETHYLPIPERAZIN-1-YL)METHYL]PHENYL}-N-[(1R)-1-PHENYLETHYL]-7H-PYRROLO[2,3-D]PYRIMIDIN-4-AMINE, EPIDERMAL GROWTH FACTOR RECEPTOR | | Authors: | Yun, C.-H, Boggon, T.J, Li, Y, Woo, S, Greulich, H, Meyerson, M, Eck, M.J. | | Deposit date: | 2006-09-29 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of Lung Cancer-Derived Egfr Mutants and Inhibitor Complexes: Mechanism of Activation and Insights Into Differential Inhibitor Sensitivity
Cancer Cell, 11, 2007
|
|
8V1F
 
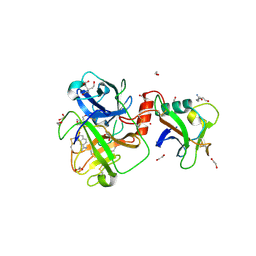 | | TMPRSS2 complexed with the noncovalent inhibitor 6-amidino-2-napthol | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fraser, B.J, Dong, A, Kutera, M, Seitova, A, Li, Y, Hutchinson, A, Edwards, A, Benard, F, Halabelian, L, Arrowsmith, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-11-20 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | TMPRSS2 complexed with the noncovalent inhibitor 6-amidino-2-napthol
To Be Published
|
|
7KA2
 
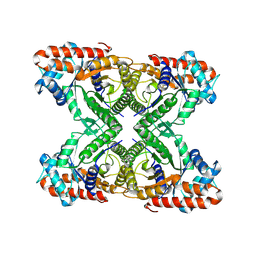 | | Aldolase, rabbit muscle (beam-tilt refinement x2) | | Descriptor: | Fructose-bisphosphate aldolase A | | Authors: | Kearns, S.K, Cash, J.N, Cianfrocco, M.A, Li, Y. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | High-resolution cryo-EM using beam-image shift at 200 keV.
Iucrj, 7, 2020
|
|
8JYZ
 
 | | Cryo-EM structure of RCD-1 pore from Neurospora crassa | | Descriptor: | Gasdermin-like protein rcd-1-1, Gasdermin-like protein rcd-1-2 | | Authors: | Hou, Y.J, Sun, Q, Li, Y, Ding, J. | | Deposit date: | 2023-07-04 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Cleavage-independent activation of ancient eukaryotic gasdermins and structural mechanisms.
Science, 384, 2024
|
|
8XXN
 
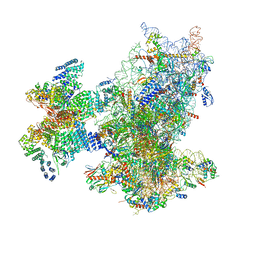 | | Cryo-EM structure of the human 43S ribosome with PDCD4 | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ye, X, Huang, Z, Li, Y, Wang, M, Cheng, J. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Human tumor suppressor PDCD4 directly interacts with ribosomes to repress translation.
Cell Res., 34, 2024
|
|
8XXL
 
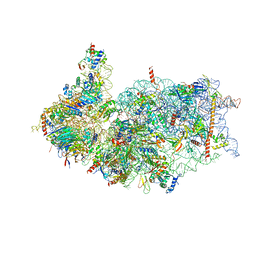 | | Cryo-EM structure of the human 40S ribosome with PDCD4 | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ye, X, Huang, Z, Li, Y, Wang, M, Cheng, J. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Human tumor suppressor PDCD4 directly interacts with ribosomes to repress translation.
Cell Res., 34, 2024
|
|
8XXM
 
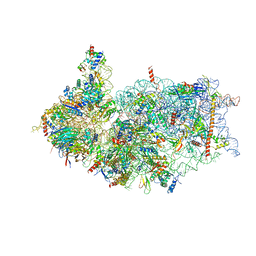 | | Cryo-EM structure of the human 40S ribosome with PDCD4 and eIF3G | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ye, X, Huang, Z, Li, Y, Wang, M, Cheng, J. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Human tumor suppressor PDCD4 directly interacts with ribosomes to repress translation.
Cell Res., 34, 2024
|
|
7KA3
 
 | | Aldolase, rabbit muscle (beam-tilt refinement x3) | | Descriptor: | Fructose-bisphosphate aldolase A | | Authors: | Kearns, S.K, Cash, J.N, Cianfrocco, M.A, Li, Y. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | High-resolution cryo-EM using beam-image shift at 200 keV.
Iucrj, 7, 2020
|
|
8H1C
 
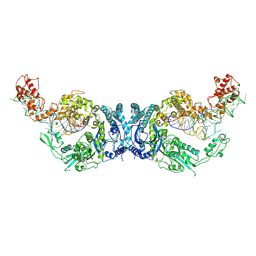 | | Cryo-EM structure of Oryza sativa plastid glycyl-tRNA synthetase in complex with two tRNAs (one in tRNA binding state and the other in tRNA locked state) | | Descriptor: | Glycine--tRNA ligase, tRNA(gly) (74-MER) | | Authors: | Yu, Z, Wu, Z, Li, Y, Lu, G, Lin, J. | | Deposit date: | 2022-10-02 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of a two-step tRNA recognition mechanism for plastid glycyl-tRNA synthetase.
Nucleic Acids Res., 51, 2023
|
|
3O73
 
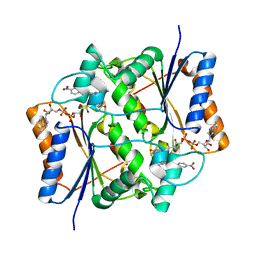 | | Crystal structure of quinone reductase 2 in complex with the indolequinone MAC627 | | Descriptor: | 5-[(4-aminobutyl)amino]-1,2-dimethyl-3-[(4-nitrophenoxy)methyl]-1H-indole-4,7-dione, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Dufour, M, Yan, C, Colucci, M.A, Siegel, D, Li, Y, De Matteis, C.I, Ross, D, Moody, C.J. | | Deposit date: | 2010-07-30 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism-Based Inhibition of Quinone Reductase 2 (NQO2): Selectivity for NQO2 over NQO1 and Structural Basis for Flavoprotein Inhibition.
Chembiochem, 12, 2011
|
|
7WGW
 
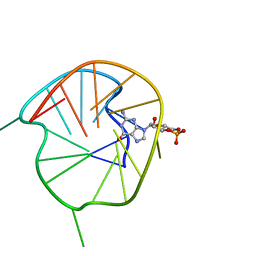 | | NMR Solution Structure of a cGMP Fill-in Vacancy G-quadruplex Formed in the Oxidized BLM Gene Promoter | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, DNA (20-MER) | | Authors: | Wang, K.B, Liu, Y, Li, Y, Li, J, Dickerhoff, J, Yang, M.H, Yang, D, Kong, L.Y. | | Deposit date: | 2021-12-29 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Oxidative Damage Induces a Vacancy G-Quadruplex That Binds Guanine Metabolites: Solution Structure of a cGMP Fill-in Vacancy G-Quadruplex in the Oxidized BLM Gene Promoter.
J.Am.Chem.Soc., 144, 2022
|
|
3O8X
 
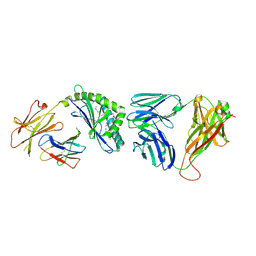 | | Recognition of Glycolipid Antigen by iNKT Cell TCR | | Descriptor: | (2S,3R)-3-HYDROXY-2-(TETRADECANOYLAMINO)OCTADECYL ALPHA-D-GALACTOPYRANOSIDURONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M, Li, Y. | | Deposit date: | 2010-08-03 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | The V alpha 14 invariant natural killer T cell TCR forces microbial glycolipids and CD1d into a conserved binding mode.
J.Exp.Med., 207, 2010
|
|
3O9W
 
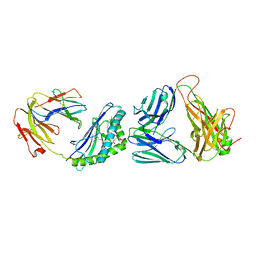 | | Recognition of a Glycolipid Antigen by the iNKT Cell TCR | | Descriptor: | (2S)-3-(alpha-D-galactopyranosyloxy)-2-(hexadecanoyloxy)propyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M, Li, Y. | | Deposit date: | 2010-08-04 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The V alpha 14 invariant natural killer T cell TCR forces microbial glycolipids and CD1d into a conserved binding mode.
J.Exp.Med., 207, 2010
|
|
2MQ6
 
 | |
5FAI
 
 | | EMG1 N1-Specific Pseudouridine Methyltransferase | | Descriptor: | CITRIC ACID, Ribosomal RNA small subunit methyltransferase NEP1, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | DONG, A, ZENG, H, LI, Y, TEMPEL, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, BROWN, P.J, WU, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-11 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | EMG1 N1-Specific Pseudouridine Methyltransferase
to be published
|
|
7K9L
 
 | | Aldolase, rabbit muscle (no beam-tilt refinement) | | Descriptor: | Fructose-bisphosphate aldolase A | | Authors: | Cianfrocco, M.A, Kearns, S.E, Cash, J.N, Li, Y. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | High-resolution cryo-EM using beam-image shift at 200 keV.
Iucrj, 7, 2020
|
|
4ERN
 
 | |
