3NR6
 
 | | Crystal structure of xenotropic murine leukemia virus-related virus (XMRV) protease | | 分子名称: | PHOSPHATE ION, POTASSIUM ION, Protease p14 | | 著者 | Lubkowski, J, Li, M, Gustchina, A, Zhou, D, Dauter, Z, Wlodawer, A. | | 登録日 | 2010-06-30 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Crystal structure of XMRV protease differs from the structures of other retropepsins.
Nat.Struct.Mol.Biol., 18, 2011
|
|
7EGQ
 
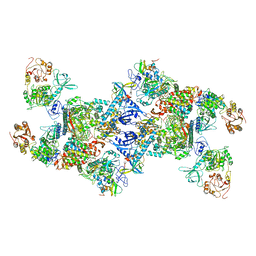 | | Co-transcriptional capping machineries in SARS-CoV-2 RTC: Coupling of N7-methyltransferase and 3'-5' exoribonuclease with polymerase reveals mechanisms for capping and proofreading | | 分子名称: | Helicase, MAGNESIUM ION, Non-structural protein 10, ... | | 著者 | Yan, L.M, Yang, Y.X, Li, M.Y, Zhang, Y, Zheng, L.T, Ge, J, Huang, Y.C, Liu, Z.Y, Wang, T, Gao, S, Zhang, R, Huang, Y.Y, Guddat, L.W, Gao, Y, Rao, Z.H, Lou, Z.Y. | | 登録日 | 2021-03-25 | | 公開日 | 2021-07-21 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.35 Å) | | 主引用文献 | Coupling of N7-methyltransferase and 3'-5' exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading.
Cell, 184, 2021
|
|
3PL9
 
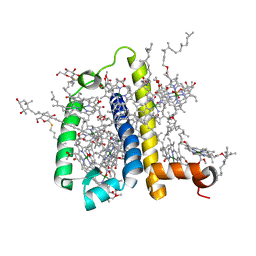 | | Crystal structure of spinach minor light-harvesting complex CP29 at 2.80 angstrom resolution | | 分子名称: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | 著者 | Pan, X.W, Li, M, Wan, T, Wang, L.F, Jia, C.J, Hou, Z.Q, Zhao, X.L, Zhang, J.P, Chang, W.R. | | 登録日 | 2010-11-14 | | 公開日 | 2011-02-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural insights into energy regulation of light-harvesting complex CP29 from spinach.
Nat.Struct.Mol.Biol., 18, 2011
|
|
6JVX
 
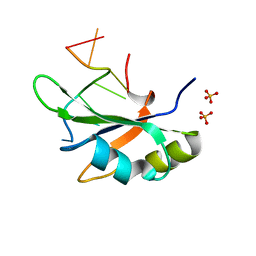 | | Crystal structure of RBM38 in complex with RNA | | 分子名称: | RNA (5'-R(*UP*GP*UP*GP*UP*GP*UP*GP*UP*GP*UP*G)-3'), RNA-binding protein 38, SULFATE ION | | 著者 | Qian, K, Li, M, Wang, J, Zhang, M, Wang, M. | | 登録日 | 2019-04-17 | | 公開日 | 2020-01-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.301 Å) | | 主引用文献 | Structural basis for mRNA recognition by human RBM38.
Biochem.J., 477, 2020
|
|
4L36
 
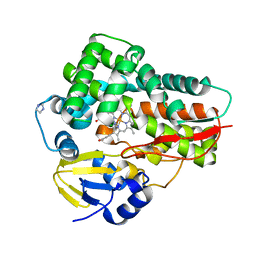 | | Crystal structure of the cytochrome P450 enzyme TxtE | | 分子名称: | HEME B/C, IMIDAZOLE, Putative P450-like protein | | 著者 | Yu, F, Li, M.J, Xu, C.Y, Wang, Z.J, Zhou, H, Yang, M, Chen, Y.X, Tang, L, He, J.H. | | 登録日 | 2013-06-05 | | 公開日 | 2013-12-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.104 Å) | | 主引用文献 | Structural Insights into the Mechanism for Recognizing Substrate of the Cytochrome P450 Enzyme TxtE.
Plos One, 8, 2013
|
|
7DZ8
 
 | | State transition supercomplex PSI-LHCI-LHCII from the LhcbM1 lacking mutant of Chlamydomonas reinhardtii | | 分子名称: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | 著者 | Pan, X.W, Li, A.J, Liu, Z.F, Li, M. | | 登録日 | 2021-01-23 | | 公開日 | 2021-06-30 | | 最終更新日 | 2021-09-01 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Structural basis of LhcbM5-mediated state transitions in green algae.
Nat.Plants, 7, 2021
|
|
7DZ7
 
 | | State transition supercomplex PSI-LHCI-LHCII from double phosphatase mutant pph1;pbcp of green alga Chlamydomonas reinhardtii | | 分子名称: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | 著者 | Pan, X.W, Li, A.J, Liu, Z.F, Li, M. | | 登録日 | 2021-01-23 | | 公開日 | 2021-06-30 | | 最終更新日 | 2021-09-01 | | 実験手法 | ELECTRON MICROSCOPY (2.84 Å) | | 主引用文献 | Structural basis of LhcbM5-mediated state transitions in green algae.
Nat.Plants, 7, 2021
|
|
7XZA
 
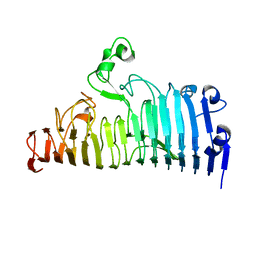 | |
7XZB
 
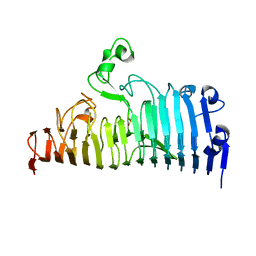 | |
7XZ8
 
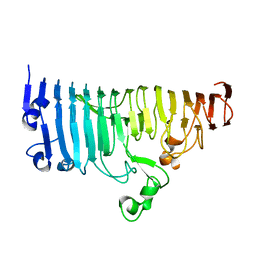 | |
7XZD
 
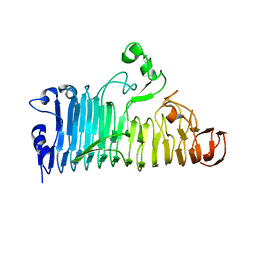 | |
6JVY
 
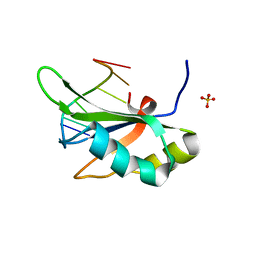 | | Crystal structure of RBM38 in complex with single-stranded DNA | | 分子名称: | DNA (5'-D(*TP*GP*TP*GP*TP*GP*TP*GP*TP*GP*TP*G)-3'), RNA-binding protein 38, SULFATE ION | | 著者 | Qian, K, Li, M, Wang, J, Zhang, M, Wang, M. | | 登録日 | 2019-04-17 | | 公開日 | 2020-01-01 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.003 Å) | | 主引用文献 | Structural basis for mRNA recognition by human RBM38.
Biochem.J., 477, 2020
|
|
6JUI
 
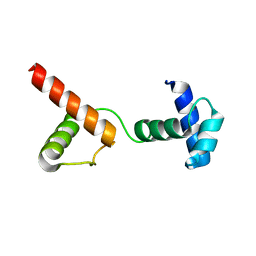 | | The atypical Myb-like protein Cdc5 contains two distinct nucleic acid-binding surfaces | | 分子名称: | Pre-mRNA-splicing factor CEF1 | | 著者 | Wang, C, Li, G, Li, M, Yang, J, Liu, J. | | 登録日 | 2019-04-14 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.402 Å) | | 主引用文献 | Two distinct nucleic acid binding surfaces of Cdc5 regulate development.
Biochem.J., 476, 2019
|
|
4RI2
 
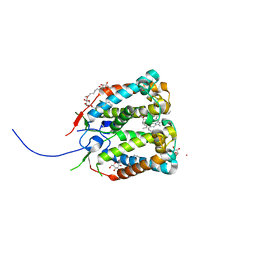 | | Crystal structure of the photoprotective protein PsbS from spinach | | 分子名称: | CHLOROPHYLL A, MERCURY (II) ION, Photosystem II 22 kDa protein, ... | | 著者 | Fan, M, Li, M, Chang, W. | | 登録日 | 2014-10-05 | | 公開日 | 2015-08-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal structures of the PsbS protein essential for photoprotection in plants.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3TPP
 
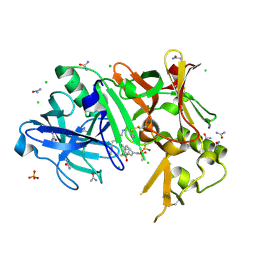 | | Crystal structure of BACE1 complexed with an inhibitor | | 分子名称: | Beta-secretase 1, CHLORIDE ION, N-[(1S,2R)-1-BENZYL-3-(CYCLOPROPYLAMINO)-2-HYDROXYPROPYL]-5-[METHYL(METHYLSULFONYL)AMINO]-N'-[(1R)-1-PHENYLETHYL]ISOPHTHALAMIDE, ... | | 著者 | Xu, Y.C, Li, M.J, Greenblatt, H, Chen, T.T, Silman, I, Sussman, J.L. | | 登録日 | 2011-09-08 | | 公開日 | 2011-11-23 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2HQG
 
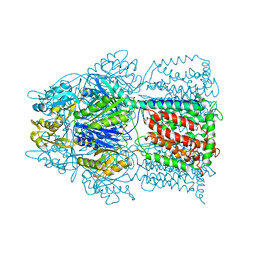 | | Conformation of the AcrB Multidrug Efflux Pump in Mutants of the Putative Proton Relay Pathway | | 分子名称: | Acriflavine resistance protein B | | 著者 | Su, C.-C, Li, M, Gu, R, Takatsuka, Y, McDermott, G, Nikaido, H, Yu, E.W. | | 登録日 | 2006-07-18 | | 公開日 | 2007-04-17 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.38 Å) | | 主引用文献 | Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway
J.Bacteriol., 188, 2006
|
|
5HTN
 
 | |
5HTJ
 
 | |
5HV7
 
 | |
5HTX
 
 | |
5HTY
 
 | |
5HTR
 
 | |
3JYN
 
 | | Crystal structures of Pseudomonas syringae pv. Tomato DC3000 quinone oxidoreductase complexed with NADPH | | 分子名称: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Quinone oxidoreductase | | 著者 | Pan, X, Zhang, H, Gao, Y, Li, M, Chang, W. | | 登録日 | 2009-09-22 | | 公開日 | 2010-01-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Crystal structures of Pseudomonas syringae pv. tomato DC3000 quinone oxidoreductase and its complex with NADPH
Biochem.Biophys.Res.Commun., 390, 2009
|
|
5HTP
 
 | |
5HU2
 
 | |
