8FZ5
 
 | | The PI31-free Bovine 20S proteasome | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Hsu, H.-C, Li, H. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | Eta igh-resolution structure of mammalian PI31-20S proteasome complex reveals mechanism of proteasome inhibition.
J.Biol.Chem., 299, 2023
|
|
6WGI
 
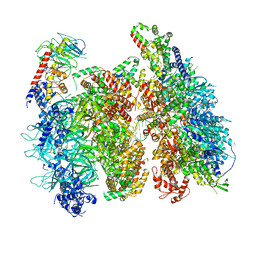 | | Atomic model of the mutant OCCM (ORC-Cdc6-Cdt1-Mcm2-7 with Mcm6 WHD truncation) loaded on DNA at 10.5 A resolution | | Descriptor: | Cell division control protein 6, Cell division cycle protein CDT1, DNA (34-MER), ... | | Authors: | Yuan, Z, Schneider, S, Dodd, T, Riera, A, Bai, L, Yan, C, Magdalou, I, Ivanov, I, Stillman, B, Li, H, Speck, C. | | Deposit date: | 2020-04-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4WN2
 
 | |
1RS9
 
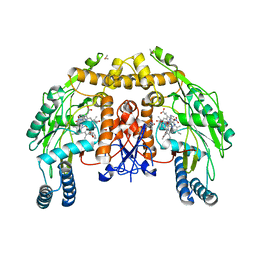 | | Bovine endothelial NOS heme domain with D-phenylalanine-D-nitroarginine amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Flinspach, M, Li, H, Jamal, J, Yang, W, Huang, H, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-12-09 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structures of the neuronal and endothelial nitric oxide synthase heme domain with D-nitroarginine-containing dipeptide inhibitors bound.
Biochemistry, 43, 2004
|
|
1RS7
 
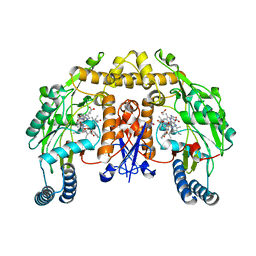 | | Rat neuronal NOS heme domain with D-phenylalanine-D-nitroarginine amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, D-MANNITOL, ... | | Authors: | Flinspach, M, Li, H, Jamal, J, Yang, W, Huang, H, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-12-09 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Neuronal and Endothelial Nitric Oxide Synthase Heme Domain with d-Nitroarginine-Containing Dipeptide Inhibitors Bound.
Biochemistry, 43, 2004
|
|
6WGG
 
 | | Atomic model of pre-insertion mutant OCCM-DNA complex(ORC-Cdc6-Cdt1-Mcm2-7 with Mcm6 WHD truncation) | | Descriptor: | Cell division control protein 6, Cell division cycle protein CDT1, DNA (41-MER), ... | | Authors: | Yuan, Z, Schneider, S, Dodd, T, Riera, A, Bai, L, Yan, C, Magdalou, I, Ivanov, I, Stillman, B, Li, H, Speck, C. | | Deposit date: | 2020-04-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6WGC
 
 | | Atomic model of semi-attached mutant OCCM-DNA complex (ORC-Cdc6-Cdt1-Mcm2-7 with Mcm6 WHD truncation) | | Descriptor: | Cell division control protein 6, DNA (41-MER), DNA replication licensing factor MCM3, ... | | Authors: | Yuan, Z, Schneider, S, Dodd, T, Riera, A, Bai, L, Yan, C, Magdalou, I, Ivanov, I, Stillman, B, Li, H, Speck, C. | | Deposit date: | 2020-04-05 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1D0C
 
 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 3-BROMO-7-NITROINDAZOLE (H4B FREE) | | Descriptor: | 3-BROMO-7-NITROINDAZOLE, ACETATE ION, BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Southan, G.J, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-09-09 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of nitric oxide synthase bound to nitro indazole reveals a novel inactivation mechanism.
Biochemistry, 40, 2001
|
|
5HJB
 
 | | AF9 YEATS in complex with histone H3 Crotonylation at K9 | | Descriptor: | Protein AF-9, peptide of Histone H3.1 | | Authors: | Li, Y.Y, Zhao, D, Guan, H.P, Li, H.T. | | Deposit date: | 2016-01-12 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain
Mol.Cell, 62, 2016
|
|
3MKA
 
 | |
5HJC
 
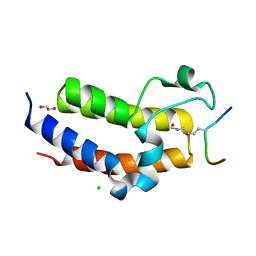 | | BRD3 second bromodomain in complex with histone H3 acetylation at K18 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 3, CHLORIDE ION, ... | | Authors: | Li, Y.Y, Zhao, D, Guan, H.P, Li, H.T. | | Deposit date: | 2016-01-13 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain
Mol.Cell, 62, 2016
|
|
1D0O
 
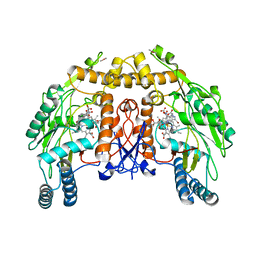 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME DOMAIN COMPLEXED WITH 3-BROMO-7-NITROINDAZOLE (H4B PRESENT) | | Descriptor: | 3-BROMO-7-NITROINDAZOLE, ACETATE ION, BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE HEME, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Southan, G.J, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1999-09-13 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of nitric oxide synthase bound to nitro indazole reveals a novel inactivation mechanism.
Biochemistry, 40, 2001
|
|
5VIE
 
 | | Electrophilic probes for deciphering substrate recognition by O-GlcNAc transferase | | Descriptor: | 2-{[(2E)-4-chlorobut-2-enoyl]amino}-2-deoxy-beta-D-glucopyranose, 2-{[(2E)-but-2-enoyl]amino}-2-deoxy-beta-D-glucopyranose, CKII, ... | | Authors: | Jiang, J, Li, B, Hu, C.-W, Worth, M, Fan, D, Li, H. | | Deposit date: | 2017-04-15 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Electrophilic probes for deciphering substrate recognition by O-GlcNAc transferase.
Nat. Chem. Biol., 13, 2017
|
|
1SN1
 
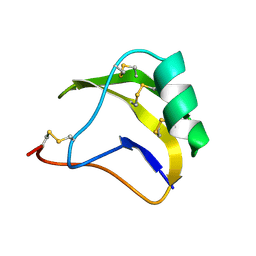 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M1 | | Descriptor: | PROTEIN (NEUROTOXIN BMK M1) | | Authors: | He, X.L, Li, H.M, Liu, X.Q, Zeng, Z.H, Wang, D.C. | | Deposit date: | 1998-11-12 | | Release date: | 1999-11-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of two alpha-like scorpion toxins: non-proline cis peptide bonds and implications for new binding site selectivity on the sodium channel.
J.Mol.Biol., 292, 1999
|
|
3V9R
 
 | | Crystal structure of Saccharomyces cerevisiae MHF complex | | Descriptor: | SULFATE ION, Uncharacterized protein YDL160C-A, Uncharacterized protein YOL086W-A | | Authors: | Yang, H, Zhang, T, Zhong, C, Li, H, Zhou, J, Ding, J. | | Deposit date: | 2011-12-28 | | Release date: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Saccharomyces Cerevisiae MHF Complex Structurally Resembles the Histones (H3-H4)(2) Heterotetramer and Functions as a Heterotetramer
Structure, 20, 2012
|
|
8PO8
 
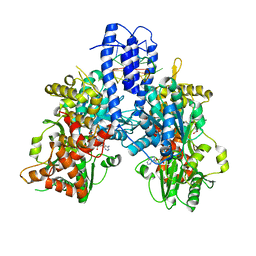 | | Structure of Escherichia coli HrpA in complex with ADP and oligonucleotide poly(dC)11 forming an i-motif | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase HrpA, DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*C)-3'), ... | | Authors: | Xin, B.G, Yuan, L.G, Zhang, L.L, Xie, S.M, Liu, N.N, Ai, X, Li, H.H, Rety, S, Xi, X.G. | | Deposit date: | 2023-07-03 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural insights into the N-terminal APHB domain of HrpA: mediating canonical and i-motif recognition.
Nucleic Acids Res., 52, 2024
|
|
8PO7
 
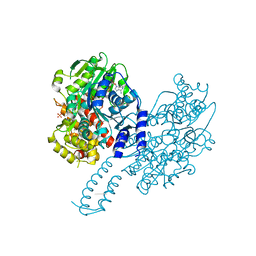 | | Structure of Escherichia coli HrpA in complex with ADP and dinucleotide dCdC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase HrpA, DNA (5'-D(P*CP*C)-3'), ... | | Authors: | Xin, B.G, Yuan, L.G, Zhang, L.L, Xie, S.M, Liu, N.N, Ai, X, Li, H.H, Rety, S, Xi, X.G. | | Deposit date: | 2023-07-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural insights into the N-terminal APHB domain of HrpA: mediating canonical and i-motif recognition.
Nucleic Acids Res., 52, 2024
|
|
2VQI
 
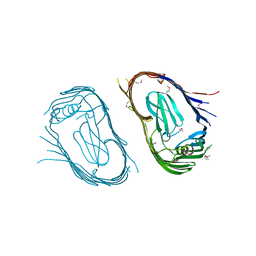 | | Structure of the P pilus usher (PapC) translocation pore | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, LAURYL DIMETHYLAMINE-N-OXIDE, OUTER MEMBRANE USHER PROTEIN PAPC | | Authors: | Remaut, H, Tang, C, Henderson, N.S, Pinkner, J.S, Wang, T, Hultgren, S.J, Thanassi, D.G, Li, H, Waksman, G. | | Deposit date: | 2008-03-16 | | Release date: | 2008-05-27 | | Last modified: | 2019-01-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Fiber formation across the bacterial outer membrane by the chaperone/usher pathway.
Cell, 133, 2008
|
|
8D1V
 
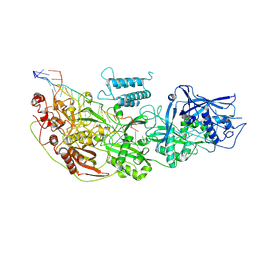 | | Cryo-EM structure of guide RNA and target RNA bound Cas7-11 | | Descriptor: | CRISPR RNA (34-MER), CRISPR-associated RAMP family protein, SS target RNA (5'-R(P*AP*GP*CP*UP*UP*GP*GP*UP*UP*CP*AP*AP*AP*GP*AP*AP*CP*G)-3'), ... | | Authors: | Rai, J, Goswami, H, Li, H. | | Deposit date: | 2022-05-27 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Molecular mechanism of active Cas7-11 in processing CRISPR RNA and interfering target RNA.
Elife, 11, 2022
|
|
4WLG
 
 | |
8D4X
 
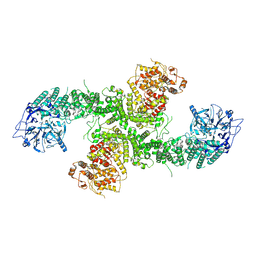 | | Structure of the human UBR5 HECT-type E3 ubiquitin ligase in a dimeric form | | Descriptor: | E3 ubiquitin-protein ligase UBR5, ZINC ION | | Authors: | Wang, F, He, Q, Lin, G, Li, H. | | Deposit date: | 2022-06-02 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the human UBR5 E3 ubiquitin ligase.
Structure, 31, 2023
|
|
4WLZ
 
 | |
3B3M
 
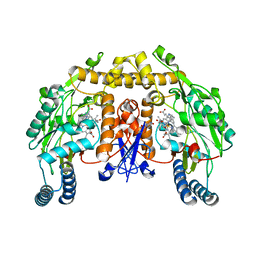 | | Structure of neuronal NOS heme domain in complex with a inhibitor (+-)-3-{cis-4'-[(6"-aminopyridin-2"-yl)methyl]pyrrolidin-3'-ylamino}propan-1-ol | | Descriptor: | 3-({(3S,4S)-4-[(6-aminopyridin-2-yl)methyl]pyrrolidin-3-yl}amino)propan-1-ol, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2007-10-22 | | Release date: | 2008-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Minimal pharmacophoric elements and fragment hopping, an approach directed at molecular diversity and isozyme selectivity. Design of selective neuronal nitric oxide synthase inhibitors.
J.Am.Chem.Soc., 130, 2008
|
|
1BVY
 
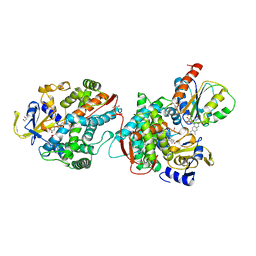 | | COMPLEX OF THE HEME AND FMN-BINDING DOMAINS OF THE CYTOCHROME P450(BM-3) | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, PROTEIN (CYTOCHROME P450 BM-3), ... | | Authors: | Sevrioukova, I.F, Li, H, Zhang, H, Peterson, J.A, Poulos, T.L. | | Deposit date: | 1998-09-21 | | Release date: | 1999-02-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of a cytochrome P450-redox partner electron-transfer complex.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
4WM0
 
 | |
