4QVZ
 
 | | FMRP N-terminal domain | | Descriptor: | 1,2-ETHANEDIOL, Fragile X mental retardation protein 1 | | Authors: | Myrick, L.K, Hashimoto, H, Cheng, X, Warren, S.T. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.195 Å) | | Cite: | Human FMRP contains an integral tandem Agenet (Tudor) and KH motif in the amino terminal domain.
Hum.Mol.Genet., 24, 2015
|
|
1JGI
 
 | |
2IA5
 
 | | T4 polynucleotide kinase/phosphatase with bound sulfate and magnesium. | | Descriptor: | ARSENIC, MAGNESIUM ION, Polynucleotide kinase, ... | | Authors: | Zhu, H, Smith, P.C, Wang, L.K, Lima, C.D, Shuman, S. | | Deposit date: | 2006-09-07 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-function analysis of the 3' phosphatase component of T4 polynucleotide kinase/phosphatase.
Virology, 366, 2007
|
|
7WQU
 
 | | FeoC from Klebsiella pneumoniae | | Descriptor: | Ferrous iron transport protein B, Probable [Fe-S]-dependent transcriptional repressor | | Authors: | Hsueh, K.L, Yu, L.K, Hsieh, Y.C, Hsiao, Y.Y, Chen, C.J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.202 Å) | | Cite: | FeoC from Klebsiella pneumoniae uses its iron sulfur cluster to regulate the GTPase activity of the ferrous iron channel.
Biochim Biophys Acta Proteins Proteom, 1871, 2023
|
|
3EHA
 
 | |
3ZHC
 
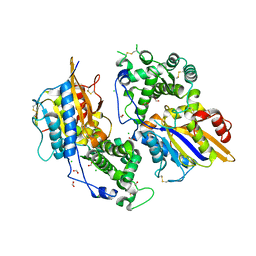 | | Structure of the phytase from Citrobacter braakii at 2.3 angstrom resolution. | | Descriptor: | CHLORIDE ION, FORMIC ACID, PHYTASE | | Authors: | Wilson, K.S, Ariza, A, Sanchez-Romero, I, Skjot, M, Vind, J, DeMaria, L, Skov, L.K, Sanchez-Ruiz, J.M. | | Deposit date: | 2012-12-20 | | Release date: | 2013-08-28 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of Protein Kinetic Stabilization by Engineered Disulfide Crosslinks
Plos One, 8, 2013
|
|
4X0N
 
 | |
6L9I
 
 | |
5NEV
 
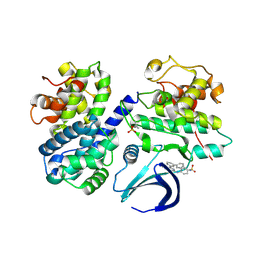 | | CDK2/Cyclin A in complex with compound 73 | | Descriptor: | 4-[[6-(3-phenylphenyl)-7~{H}-purin-2-yl]amino]benzenesulfonamide, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Coxon, C.R, Anscombe, E, Harnor, S.J, Martin, M.P, Carbain, B, Hardcastle, I.R, Harlow, L.K, Korolchuk, S, Matheson, C.J, Noble, M.E.M, Newell, D.R, Turner, D, Sivaprakasam, M, Wang, L.Z, Wong, C, Golding, B.T, Griffin, R.J, Cano, G. | | Deposit date: | 2017-03-12 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Cyclin-Dependent Kinase (CDK) Inhibitors: Structure-Activity Relationships and Insights into the CDK-2 Selectivity of 6-Substituted 2-Arylaminopurines.
J. Med. Chem., 60, 2017
|
|
4W93
 
 | | Human pancreatic alpha-amylase in complex with montbretin A | | Descriptor: | CALCIUM ION, CHLORIDE ION, Montbretin A, ... | | Authors: | Williams, L.K, Caner, S, Brayer, G.D. | | Deposit date: | 2014-08-27 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.352 Å) | | Cite: | The amylase inhibitor montbretin A reveals a new glycosidase inhibition motif.
Nat.Chem.Biol., 11, 2015
|
|
8FR5
 
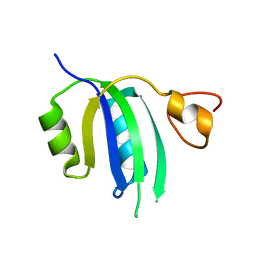 | | Crystal structure of the Human Smacovirus 1 Rep domain | | Descriptor: | MANGANESE (II) ION, Rep, SODIUM ION | | Authors: | Limon, L.K, Shi, K, Dao, A, Rugloski, J, Tompkins, K.J, Aihara, H, Gordon, W.R, Evans IIII, R.L. | | Deposit date: | 2023-01-06 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | The crystal structure of the human smacovirus 1 Rep domain.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
7K7P
 
 | |
6NCA
 
 | | HLA-A2 (A*02:01) bound to a peptide from the Epstein-Barr virus BRLF1 protein | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Stern, L.J, Selin, L.K, Song, I.Y. | | Deposit date: | 2018-12-11 | | Release date: | 2019-10-23 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (3.300001 Å) | | Cite: | CDR3 alpha drives selection of the immunodominant Epstein Barr virus (EBV) BRLF1-specific CD8 T cell receptor repertoire in primary infection.
Plos Pathog., 15, 2019
|
|
9BNR
 
 | | 4-(2-isothiocyanatoethyl)benzenesulfonamide complexed with Macrophage Migration Inhibitory Factor | | Descriptor: | Macrophage migration inhibitory factor, N-[2-(4-sulfamoylphenyl)ethyl]methanethioamide, SULFATE ION | | Authors: | Fellner, M, Rutledge, M.T, Putha, L, Kok, L.K, Gamble, A.B, Wilbanks, S.M, Vernall, A.J, Tyndall, J.D.A. | | Deposit date: | 2024-05-02 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Covalent isothiocyanate inhibitors of macrophage migration inhibitory factor as potential colorectal cancer treatments.
Chemmedchem, 2024
|
|
9BNQ
 
 | | N-(4-(isothiocyanatomethyl)phenyl)methanesulfonamide complexed with Macrophage Migration Inhibitory Factor | | Descriptor: | ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor, N-{[4-(methanesulfonamido)phenyl]methyl}methanethioamide, ... | | Authors: | Fellner, M, Rutledge, M.T, Putha, L, Kok, L.K, Gamble, A.B, Wilbanks, S.M, Vernall, A.J, Tyndall, J.D.A. | | Deposit date: | 2024-05-02 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Covalent isothiocyanate inhibitors of macrophage migration inhibitory factor as potential colorectal cancer treatments.
Chemmedchem, 2024
|
|
5LAR
 
 | | Crystal structure of p38 alpha MAPK14 in complex with VPC00628 | | Descriptor: | 5-azanyl-~{N}-[[4-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Petersen, L.K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel p38alpha MAP kinase inhibitors identified from yoctoReactor DNA-encoded small molecule library
Medchemcomm, 7, 2016
|
|
6WDQ
 
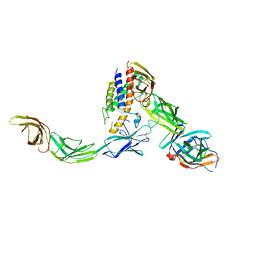 | | IL23/IL23R/IL12Rb1 signaling complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1, ... | | Authors: | Jude, K.M, Ely, L.K, Glassman, C.R, Thomas, C, Spangler, J.B, Lupardus, P.J, Garcia, K.C. | | Deposit date: | 2020-04-01 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for IL-12 and IL-23 receptor sharing reveals a gateway for shaping actions on T versus NK cells.
Cell, 184, 2021
|
|
6X1K
 
 | | Solution NMR structure of de novo designed TMB2.3 | | Descriptor: | De novo designed transmembrane beta-barrel TMB2.3 | | Authors: | Liang, B, Vorobieva, A.A, Chow, C.M, Baker, D, Tamm, L.K. | | Deposit date: | 2020-05-19 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo design of transmembrane beta barrels.
Science, 371, 2021
|
|
5LQF
 
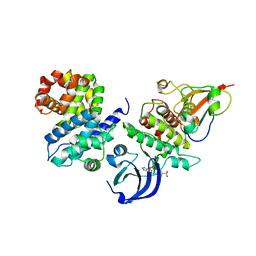 | | CDK1/CyclinB1/CKS2 in complex with NU6102 | | Descriptor: | Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, G2/mitotic-specific cyclin-B1, ... | | Authors: | Coxon, C.R, Anscombe, E, Harnor, S.J, Martin, M.P, Carbain, B.J, Hardcastle, I.R, Harlow, L.K, Korolchuk, S, Matheson, C.J, Noble, M.E, Newell, D.R, Turner, D.M, Sivaprakasam, M, Wang, L.Z, Wong, C, Golding, B.T, Griffin, R.J, Endicott, J.A, Cano, C. | | Deposit date: | 2016-08-17 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Cyclin-Dependent Kinase (CDK) Inhibitors: Structure-Activity Relationships and Insights into the CDK-2 Selectivity of 6-Substituted 2-Arylaminopurines.
J. Med. Chem., 60, 2017
|
|
6ZM8
 
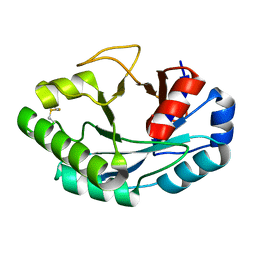 | | Structure of muramidase from Acremonium alcalophilum | | Descriptor: | muramidase | | Authors: | Moroz, O.V, Blagova, E, Taylor, E, Turkenburg, J.P, Skov, L.K, Gippert, G.P, Schnorr, K.M, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Nymand-Grarup, S, Davies, G.J, Wilson, K.S. | | Deposit date: | 2020-07-01 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.78 Å) | | Cite: | Fungal GH25 muramidases: New family members with applications in animal nutrition and a crystal structure at 0.78 angstrom resolution.
Plos One, 16, 2021
|
|
6ZMV
 
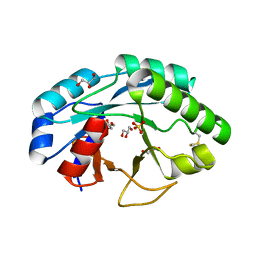 | | Structure of muramidase from Trichobolus zukalii | | Descriptor: | GLYCEROL, SULFATE ION, muramidase | | Authors: | Moroz, O.V, Blagova, E, Taylor, E, Turkenburg, J.P, Skov, L.K, Gippert, G.P, Schnorr, K.M, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Nymand-Grarup, S, Davies, G.J, Wilson, K.S. | | Deposit date: | 2020-07-04 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Fungal GH25 muramidases: New family members with applications in animal nutrition and a crystal structure at 0.78 angstrom resolution.
Plos One, 16, 2021
|
|
6OQH
 
 | |
6T5S
 
 | | Apo form of C-type lysozyme from the upper gastrointestinal tract of Opisthocomus hoatzin | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C | | Authors: | Taylor, E.J, Skjot, M, Skov, L.K, Klausen, M, De Maria, L, Gippert, G.P, Turkenburg, J.P, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-10-17 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The C-Type Lysozyme from the upper Gastrointestinal Tract of Opisthocomus hoatzin, the Stinkbird.
Int J Mol Sci, 20, 2019
|
|
6TGK
 
 | | Domain swapped E6AP C-lobe dimer | | Descriptor: | ACETATE ION, CALCIUM ION, Ubiquitin-protein ligase E3A | | Authors: | Ries, L.K, Feiler, C, Lowe, L.D, Liess, A.K.L, Lorenz, S. | | Deposit date: | 2019-11-16 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the catalytic C-lobe of the HECT-type ubiquitin ligase E6AP.
Protein Sci., 29, 2020
|
|
2WPG
 
 | | Sucrose Hydrolase | | Descriptor: | AMYLOSUCRASE OR ALPHA AMYLASE | | Authors: | Champion, E, Remaud-Simeon, M, Skov, L.K, Kastrup, J.S, Gajhede, M, Mirza, O. | | Deposit date: | 2009-08-06 | | Release date: | 2009-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Apo Structure of Sucrose Hydrolase from Xanthomonas Campestris Pv. Campestris Shows an Open Active-Site Groove
Acta Crystallogr.,Sect.D, 65, 2009
|
|
