1M7A
 
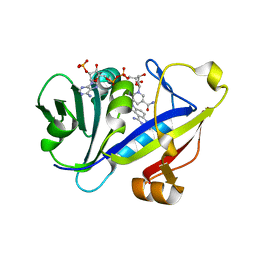 | | CANDIDA ALBICANS DIHYDROFOLATE REDUCTASE COMPLEXED WITH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE (NADPH) AND 7-[2-methoxy-1-(methoxymethyl)ethyl]-7H-pyrrolo[3,2-f] quinazoline-1,3-diamine (GW557) | | Descriptor: | 7-[2-METHOXY-1-(METHOXYMETHYL)ETHYL]-7H-PYRROLO[3,2-F] QUINAZOLINE-1,3-DIAMINE, DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Whitlow, M, Howard, A.J, Kuyper, L.F. | | Deposit date: | 2002-07-19 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | X-Ray Crystallographic Studies of Candida Albicans Dihydrofolate Reductase. High Resolution Structures of the Holoenzyme and an Inhibited Ternary Complex.
J.Biol.Chem., 272, 1997
|
|
6ZFN
 
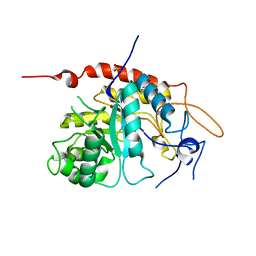 | | Structure of an inactive E404Q variant of the catalytic domain of human endo-alpha-mannosidase MANEA in complex with 1-methyl alpha-1,2-mannobiose | | Descriptor: | Glycoprotein endo-alpha-1,2-mannosidase, SULFATE ION, alpha-D-mannopyranose-(1-2)-methyl alpha-D-mannopyranoside | | Authors: | Sobala, L.F, Fernandes, P.Z, Hakki, Z, Thompson, A.J, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5MEL
 
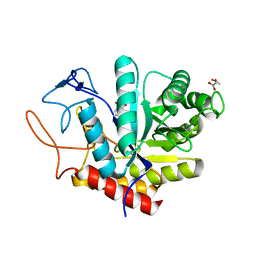 | | Structure of an E333Q variant of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with Glc-alpha-1,3-(3R,4R,5R)-5-(hydroxymethyl)cyclohex-1,2-ene-3,4-diol | | Descriptor: | (1~{R},2~{R},6~{R})-6-(hydroxymethyl)cyclohex-3-ene-1,2-diol, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Petricevic, M, Sobala, L.F, Fernandes, P.Z, Raich, L, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-11-15 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Contribution of Shape and Charge to the Inhibition of a Family GH99 endo-alpha-1,2-Mannanase.
J. Am. Chem. Soc., 139, 2017
|
|
2JQQ
 
 | | Solution structure of Saccharomyces cerevisiae conserved oligomeric Golgi subunit 2 protein (Cog2p) | | Descriptor: | Conserved oligomeric Golgi complex subunit 2 | | Authors: | Cavanaugh, L.F, Chen, X, Pelczer, I, Rizo, J, Hughson, F.M. | | Deposit date: | 2007-06-06 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of conserved oligomeric Golgi complex subunit 2
J.Biol.Chem., 282, 2007
|
|
4L5G
 
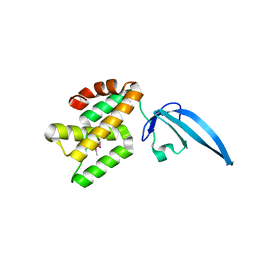 | | Crystal structure of Thermus thermophilus CarD | | Descriptor: | CarD | | Authors: | Srivastava, D.B, Westblade, L.F, Campbell, E.A, Darst, S.A. | | Deposit date: | 2013-06-11 | | Release date: | 2013-07-31 | | Last modified: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (2.3902 Å) | | Cite: | Structure and function of CarD, an essential mycobacterial transcription factor.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5MU5
 
 | | Structure of MAf glycosyltransferase from Magnetospirillum magneticum AMB-1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Murat, D, Vincentelli, R, Wu, L.F, Guerardel, Y, Alberto, F. | | Deposit date: | 2017-01-12 | | Release date: | 2017-11-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Glycosylate and move! The glycosyltransferase Maf is involved in bacterial flagella formation.
Environ. Microbiol., 20, 2018
|
|
2N34
 
 | | NMR assignments and solution structure of the JAK interaction region of SOCS5 | | Descriptor: | Suppressor of cytokine signaling 5 | | Authors: | Chandrashekaran, I.R, Mohanty, B, Linossi, E.M, Nicholson, S.E, Babon, J, Norton, R.S, Dagley, L.F, Leung, E.W.W, Murphy, J.M. | | Deposit date: | 2015-05-21 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Functional Characterization of the Conserved JAK Interaction Region in the Intrinsically Disordered N-Terminus of SOCS5.
Biochemistry, 54, 2015
|
|
4LU9
 
 | | Crystal structure of E.coli SbcD at 2.5 angstrom resolution | | Descriptor: | Exonuclease subunit SbcD, GLYCEROL | | Authors: | Liu, S, Tian, L.F, Yan, X.X, Liang, D.C. | | Deposit date: | 2013-07-25 | | Release date: | 2014-08-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for DNA recognition and nuclease processing by the Mre11 homologue SbcD in double-strand breaks repair.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4XH3
 
 | |
4XHK
 
 | | PIM1 kinase in complex with Compound 1s | | Descriptor: | 2-(2,6-difluorophenyl)-N-{4-[(3S)-pyrrolidin-3-yloxy]pyridin-3-yl}-1,3-thiazole-4-carboxamide, Serine/threonine-protein kinase pim-1 | | Authors: | Marcotte, D.J, Silvian, L.F. | | Deposit date: | 2015-01-05 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based design of low-nanomolar PIM kinase inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
1UOD
 
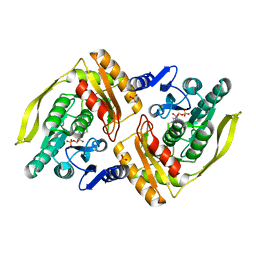 | | Crystal structure of the dihydroxyacetone kinase from E. coli in complex with dihydroxyacetone-phosphate | | Descriptor: | DIHYDROXYACETONE KINASE, GLYCERALDEHYDE-3-PHOSPHATE, SULFATE ION | | Authors: | Siebold, C, Garcia-Alles, L.F, Luthi-Nyffeler, T, Flukiger-Bruhwiler, K, Burgi, H.-B, Baumann, U, Erni, B. | | Deposit date: | 2003-09-16 | | Release date: | 2004-09-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phosphoenolpyruvate- and ATP-Dependent Dihydroxyacetone Kinases: Covalent Substrate-Binding and Kinetic Mechanism
Biochemistry, 43, 2004
|
|
1KC6
 
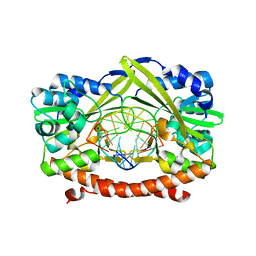 | | HincII Bound to Cognate DNA | | Descriptor: | 5'-D(P*CP*CP*GP*GP*TP*CP*GP*AP*CP*CP*GP*G)-3', SODIUM ION, TYPE II RESTRICTION ENZYME HINCII | | Authors: | Horton, N.C, Dorner, L.F, Perona, J.J. | | Deposit date: | 2001-11-07 | | Release date: | 2001-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sequence selectivity and degeneracy of a restriction endonuclease mediated by DNA intercalation.
Nat.Struct.Biol., 9, 2002
|
|
5GRM
 
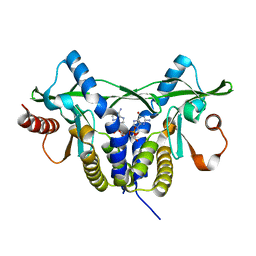 | | Crystal structure of rat STING in complex with cyclic GMP-AMP with 2'5'and 3'5'phosphodiester linkage(2'3'-cGAMP) | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Zhang, H, Han, M.J, Tao, J.L, Ye, Z.Y, Du, X.X, Deng, M.J, Zhang, X.Y, Li, L.F, Jiang, Z.F, Su, X.D. | | Deposit date: | 2016-08-11 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of rat STING in complex with cyclic GMP-AMP with 2'5'and 3'5'phosphodiester linkage(2'3'-cGAMP)
To Be Published
|
|
1KE8
 
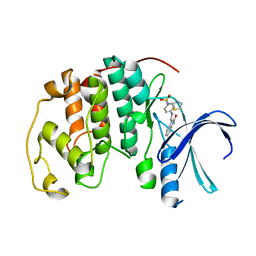 | | CYCLIN-DEPENDENT KINASE 2 (CDK2) COMPLEXED WITH 4-{[(2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]AMINO}-N-(1,3-THIAZOL-2-YL)BENZENESULFONAMIDE | | Descriptor: | 4-{[(2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]AMINO}-N-(1,3-THIAZOL-2-YL)BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.M, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
1KE6
 
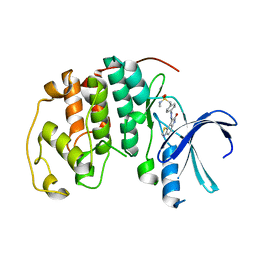 | | CYCLIN-DEPENDENT KINASE 2 (CDK2) COMPLEXED WITH N-METHYL-{4-[2-(7-OXO-6,7-DIHYDRO-8H-[1,3]THIAZOLO[5,4-E]INDOL-8-YLIDENE)HYDRAZINO]PHENYL}METHANESULFONAMIDE | | Descriptor: | Cell division protein kinase 2, N-METHYL-{4-[2-(7-OXO-6,7-DIHYDRO-8H-[1,3]THIAZOLO[5,4-E]INDOL-8-YLIDENE)HYDRAZINO]PHENYL}METHANESULFONAMIDE | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.H, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
1KE9
 
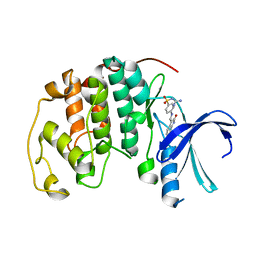 | | CYCLIN-DEPENDENT KINASE 2 (CDK2) COMPLEXED WITH 3-{[4-({[AMINO(IMINO)METHYL]AMINOSULFONYL)ANILINO]METHYLENE}-2-OXO-2,3-DIHYDRO-1H-INDOLE | | Descriptor: | 3-{[4-([AMINO(IMINO)METHYL]AMINOSULFONYL)ANILINO]METHYLENE}-2-OXO-2,3-DIHYDRO-1H-INDOLE, Cell division protein kinase 2 | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.M, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
1KE7
 
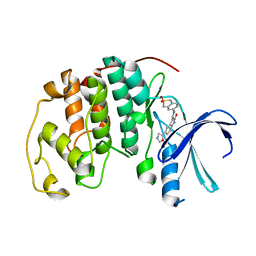 | | CYCLIN-DEPENDENT KINASE 2 (CDK2) COMPLEXED WITH 3-{[(2,2-DIOXIDO-1,3-DIHYDRO-2-BENZOTHIEN-5-YL)AMINO]METHYLENE}-5-(1,3-OXAZOL-5-YL)-1,3-DIHYDRO-2H-INDOL-2-ONE | | Descriptor: | 3-{[(2,2-DIOXIDO-1,3-DIHYDRO-2-BENZOTHIEN-5-YL)AMINO]METHYLENE}-5-(1,3-OXAZOL-5-YL)-1,3-DIHYDRO-2H-INDOL-2-ONE, Cell division protein kinase 2 | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.H, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
1KE5
 
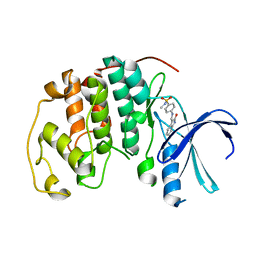 | | CDK2 complexed with N-methyl-4-{[(2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]amino}benzenesulfonamide | | Descriptor: | Cell division protein kinase 2, N-METHYL-4-{[(2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]AMINO}BENZENESULFONAMIDE | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.M, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
5GS5
 
 | | Crystal structure of apo rat STING | | Descriptor: | SULFATE ION, Stimulator of interferon genes protein | | Authors: | Zhang, H, Han, M.J, Tao, J.L, Ye, Z.Y, Du, X.X, Deng, M.J, Zhang, X.Y, Li, L.F, Jiang, Z.F, Su, X.D. | | Deposit date: | 2016-08-13 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of apo ratSTING
To Be Published
|
|
5GY3
 
 | |
5HYS
 
 | | Structure of IgE complexed with omalizumab | | Descriptor: | Epididymis luminal protein 214, Ig epsilon chain C region, SULFATE ION, ... | | Authors: | Pennington, L.F, Tarchevskaya, S.S, Sathiyamoorthy, K, Jardetzky, T.S. | | Deposit date: | 2016-02-01 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of omalizumab therapy and omalizumab-mediated IgE exchange.
Nat Commun, 7, 2016
|
|
1XA6
 
 | | Crystal Structure of the Human Beta2-Chimaerin | | Descriptor: | Beta2-chimaerin, ZINC ION | | Authors: | Canagarajah, B, Leskow, F.C, Ho, J.Y, Mischak, H, Saidi, L.F, Kazanietz, M.G, Hurley, J.H. | | Deposit date: | 2004-08-25 | | Release date: | 2004-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural mechanism for lipid activation of the Rac-specific GAP, beta2-chimaerin.
Cell(Cambridge,Mass.), 119, 2004
|
|
5FVD
 
 | | Human metapneumovirus N0-P complex | | Descriptor: | CHLORIDE ION, NUCLEOCAPSID, PHOSPHOPROTEIN | | Authors: | Renner, M, Bertinelli, M, Leyrat, C, Paesen, G.C, Saraiva de Oliveira, L.F, Huiskonen, J.T, Grimes, J.M. | | Deposit date: | 2016-02-05 | | Release date: | 2016-02-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Nucleocapsid assembly in pneumoviruses is regulated by conformational switching of the N protein.
Elife, 5, 2016
|
|
5AC2
 
 | | human aldehyde dehydrogenase 1A1 with duocarmycin analog | | Descriptor: | 1-[(1S)-1-methyl-5-oxidanyl-1,2-dihydrobenzo[e]indol-3-yl]hexan-1-one, RETINAL DEHYDROGENASE 1, YTTERBIUM (III) ION, ... | | Authors: | Koch, M.F, Harteis, S, Blank, I.D, Pestel, G, Tietze, L.F, Ochsenfeld, C, Schneider, S, Sieber, S.A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural, Biochemical, and Computational Studies Reveal the Mechanism of Selective Aldehyde Dehydrogenase 1A1 Inhibition by Cytotoxic Duocarmycin Analogues.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7KGK
 
 | | Crystal structure of synthetic nanobody (Sb16) complexes with SARS-CoV-2 receptor binding domain | | Descriptor: | Sb16, Sybody-16, Synthetic Nanobody, ... | | Authors: | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2020-10-16 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
