3OB4
 
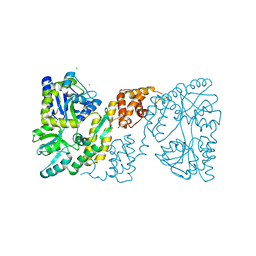 | | MBP-fusion protein of the major peanut allergen Ara h 2 | | Descriptor: | CHLORIDE ION, Maltose ABC transporter periplasmic protein,Arah 2, SULFATE ION, ... | | Authors: | Mueller, G.A, Gosavi, R.A, Moon, A.F, London, R.E, Pedersen, L.C. | | Deposit date: | 2010-08-06 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | Ara h 2: crystal structure and IgE binding distinguish two subpopulations of peanut allergic patients by epitope diversity.
Allergy, 66, 2011
|
|
3OAI
 
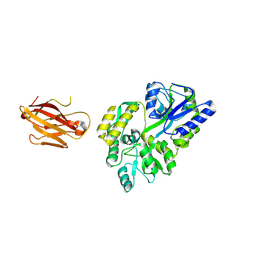 | | Crystal structure of the extra-cellular domain of human myelin protein zero | | Descriptor: | Maltose-binding periplasmic protein, Myelin protein P0, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Sohi, J, Kamholz, J, Kovari, L.C. | | Deposit date: | 2010-08-05 | | Release date: | 2011-12-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the extracellular domain of human myelin protein zero.
Proteins, 80, 2012
|
|
3OQ7
 
 | | Crystal Structures of Multidrug-Resistant Clinical Isolate 769 HIV-1 Protease Variants | | Descriptor: | HIV-1 Protease | | Authors: | Yedidi, R.S, Proteasa, G, Martinez-Cajas, J.L, Vickrey, J.F, Martin, P.D, Wawrzak, Z, Kovari, L.C. | | Deposit date: | 2010-09-02 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Contribution of the 80s loop of HIV-1 protease to the multidrug-resistance mechanism: crystallographic study of MDR769 HIV-1 protease variants.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3OQA
 
 | | Crystal Structures of Multidrug-Resistant Clinical Isolate 769 HIV-1 Protease Variants | | Descriptor: | HIV-1 Protease | | Authors: | Yedidi, R.S, Proteasa, G, Martinez-Cajas, J.L, Vickrey, J.F, Martin, P.D, Wawrzak, Z, Kovari, L.C. | | Deposit date: | 2010-09-02 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Contribution of the 80s loop of HIV-1 protease to the multidrug-resistance mechanism: crystallographic study of MDR769 HIV-1 protease variants.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3OU1
 
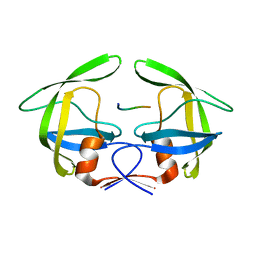 | | MDR769 HIV-1 protease complexed with RH/IN hepta-peptide | | Descriptor: | MDR HIV-1 protease, RH/IN substrate peptide | | Authors: | Liu, Z, Wang, Y, Brunzelle, J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2010-09-14 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Nine Crystal Structures Determine the Substrate Envelope of the MDR HIV-1 Protease.
Protein J., 30, 2011
|
|
2VOK
 
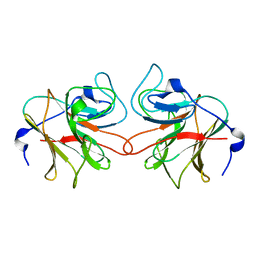 | | Murine TRIM21 | | Descriptor: | 52 KDA RO PROTEIN | | Authors: | Keeble, A.H, Khan, Z, Forster, A, James, L.C. | | Deposit date: | 2008-02-19 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Trim21 is an Igg Receptor that is Structurally, Thermodynamically, and Kinetically Conserved.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4JRB
 
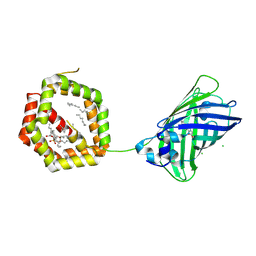 | | Structure of Cockroach Allergen Bla g 1 Tandem Repeat as a EGFP fusion | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, CHLORIDE ION, DODECANE, ... | | Authors: | Mueller, G.A, Pedersen, L.C, Lih, F.B, Glesner, J, Moon, A.F, Chapman, M.D, Tomer, K, London, R.E. | | Deposit date: | 2013-03-21 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.414 Å) | | Cite: | The novel structure of the cockroach allergen Bla g 1 has implications for allergenicity and exposure assessment.
J.Allergy Clin.Immunol., 132, 2013
|
|
2X83
 
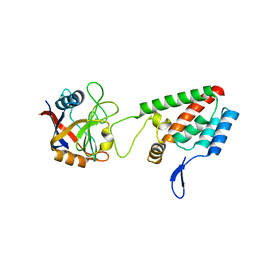 | |
2X2A
 
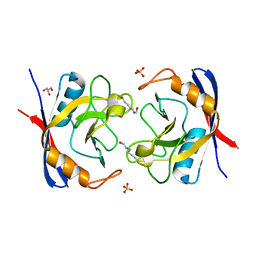 | | Free acetyl-CypA trigonal form | | Descriptor: | GLYCEROL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A, SULFATE ION | | Authors: | Lammers, M, Neumann, H, Chin, J.W, James, L.C. | | Deposit date: | 2010-01-12 | | Release date: | 2010-03-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Acetylation Regulates Cyclophilin a Catalysis, Immunosuppression and HIV Isomerization.
Nat.Chem.Biol., 6, 2010
|
|
2X2D
 
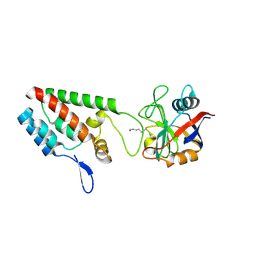 | | acetyl-CypA:HIV-1 N-term capsid domain complex | | Descriptor: | CAPSID PROTEIN P24, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Lammers, M, Neumann, H, Chin, J.W, James, L.C. | | Deposit date: | 2010-01-12 | | Release date: | 2010-03-23 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Acetylation Regulates Cyclophilin a Catalysis, Immunosuppression and HIV Isomerisation
Nat.Chem.Biol., 6, 2010
|
|
2YOK
 
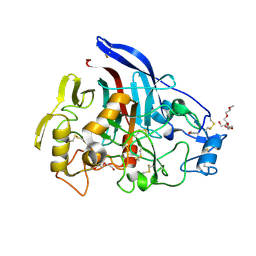 | | Cellobiohydrolase I Cel7A from Trichoderma harzianum at 1.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Textor, L.C, Colussi, F, Serpa, V, Squina, F.M, Pereira Jr, N, Polikarpov, I. | | Deposit date: | 2012-10-25 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Joint X-Ray Crystallographic and Molecular Dynamics Study of Cellobiohydrolase I from Trichoderma Harzianum: Deciphering the Structural Features of Cellobiohydrolase Catalytic Activity.
FEBS J., 280, 2013
|
|
3OQD
 
 | | Crystal Structures of Multidrug-Resistant Clinical Isolate 769 HIV-1 Protease Variants | | Descriptor: | HIV-1 Protease | | Authors: | Yedidi, R.S, Proteasa, G, Martinez-Cajas, J.L, Vickrey, J.F, Martin, P.D, Wawrzak, Z, Kovari, L.C. | | Deposit date: | 2010-09-02 | | Release date: | 2011-04-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Contribution of the 80s loop of HIV-1 protease to the multidrug-resistance mechanism: crystallographic study of MDR769 HIV-1 protease variants.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2WOZ
 
 | | The novel beta-propeller of the BTB-Kelch protein Krp1 provides the binding site for Lasp-1 that is necessary for pseudopodia extension | | Descriptor: | KELCH REPEAT AND BTB DOMAIN-CONTAINING PROTEIN 10 | | Authors: | Gray, C.H, McGarry, L.C, Spence, H.J, Riboldi-Tunnicliffe, A, Ozanne, B.W. | | Deposit date: | 2009-07-31 | | Release date: | 2009-09-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel beta-propeller of the BTB-Kelch protein Krp1 provides a binding site for Lasp-1 that is necessary for pseudopodial extension.
J. Biol. Chem., 284, 2009
|
|
2M5S
 
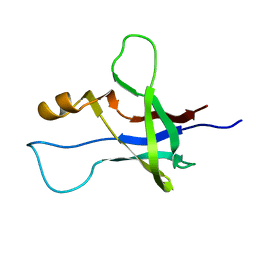 | | High-resolution NMR structure and cryo-EM imaging support multiple functional roles for the accessory I-domain of phage P22 coat protein | | Descriptor: | Coat protein | | Authors: | Rizzo, A.A, Suhanovsky, M.M, Baker, M.L, Fraser, L.C.R, Jones, L.M, Rempel, D.L, Gross, M.L, Chiu, W, Alexandrescu, A.T, Teschke, C.M. | | Deposit date: | 2013-03-05 | | Release date: | 2014-03-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Multiple Functional Roles of the Accessory I-Domain of Bacteriophage P22 Coat Protein Revealed by NMR Structure and CryoEM Modeling.
Structure, 22, 2014
|
|
2WJN
 
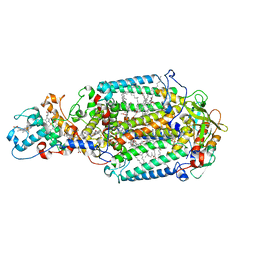 | | Lipidic sponge phase crystal structure of photosynthetic reaction centre from Blastochloris viridis (high dose) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Wohri, A.B, Wahlgren, W.Y, Malmerberg, E, Johansson, L.C, Neutze, R, Katona, G. | | Deposit date: | 2009-05-27 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Lipidic sponge phase crystal structure of a photosynthetic reaction center reveals lipids on the protein surface.
Biochemistry, 48, 2009
|
|
2X2C
 
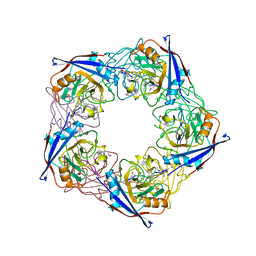 | | acetyl-CypA:cyclosporine complex | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Lammers, M, Neumann, H, Chin, J.W, James, L.C. | | Deposit date: | 2010-01-12 | | Release date: | 2010-03-23 | | Last modified: | 2018-05-02 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Acetylation Regulates Cyclophilin a Catalysis, Immunosuppression and HIV Isomerisation
Nat.Chem.Biol., 6, 2010
|
|
2X5U
 
 | | 80 microsecond Laue diffraction snapshot from crystals of a photosynthetic reaction centre without illumination. | | Descriptor: | BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, FE (II) ION, ... | | Authors: | Wohri, A.B, Katona, G, Johansson, L.C, Fritz, E, Malmerberg, E, Andersson, M, Vincent, J, Eklund, M, Cammarata, M, Wulff, M, Davidsson, J, Groenhof, G, Neutze, R. | | Deposit date: | 2010-02-10 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Light-induced structural changes in a photosynthetic reaction center caught by Laue diffraction.
Science, 328, 2010
|
|
4KHU
 
 | |
4KHW
 
 | |
4KI6
 
 | |
4KHY
 
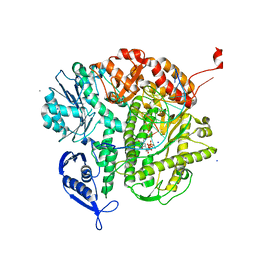 | |
4KI4
 
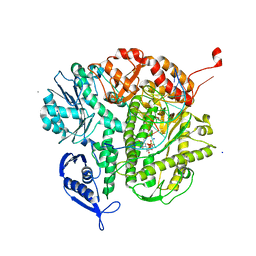 | |
4KKD
 
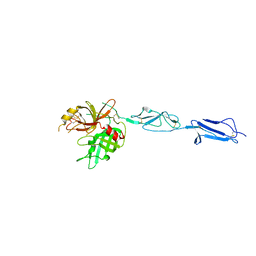 | | The X-ray crystal structure of Mannose-binding lectin-associated serine proteinase-3 reveals the structural basis for enzyme inactivity associated with the 3MC syndrome | | Descriptor: | IMIDAZOLE, Mannan-binding lectin serine protease 1 | | Authors: | Yongqing, T, Wilmann, P.G, Reeve, S.B, Coetzer, T.H, Smith, A.I, Whisstock, J.C, Pike, R.N, Wijeyewickrema, L.C. | | Deposit date: | 2013-05-05 | | Release date: | 2013-07-03 | | Last modified: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (2.5991 Å) | | Cite: | The X-ray Crystal Structure of Mannose-binding Lectin-associated Serine Proteinase-3 Reveals the Structural Basis for Enzyme Inactivity Associated with the Carnevale, Mingarelli, Malpuech, and Michels (3MC) Syndrome.
J.Biol.Chem., 288, 2013
|
|
2ICT
 
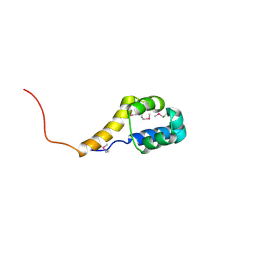 | | Crystal structure of the bacterial antitoxin HigA from Escherichia coli at pH 8.5. Northeast Structural Genomics TARGET ER390. | | Descriptor: | antitoxin higa | | Authors: | Arbing, M.A, Abashidze, M, Hurley, J.M, Zhao, L, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Inouye, M, Woychik, N.A, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-09-26 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structures of Phd-Doc, HigA, and YeeU Establish Multiple Evolutionary Links between Microbial Growth-Regulating Toxin-Antitoxin Systems.
Structure, 18, 2010
|
|
2ID1
 
 | | X-Ray Crystal Structure of Protein CV0518 from Chromobacterium violaceum, Northeast Structural Genomics Consortium Target CvR5. | | Descriptor: | Hypothetical protein, IODIDE ION | | Authors: | Forouhar, F, Zhou, W, Seetharaman, J, Ho, C.K, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-13 | | Release date: | 2006-09-26 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: |
|
|
