6G1M
 
 | | Amine Dehydrogenase from Petrotoga mobilis; open and closed form | | Descriptor: | Dihydrodipicolinate reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Beloti, L, Frese, A, Mayol, O, Vaxelaire-Vergne, C, Grogan, G. | | Deposit date: | 2018-03-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A family of native amine dehydrogenases for the asymmetric reductive amination of ketones
Nat Catal, 2019
|
|
6G1Z
 
 | | Crystal structure of a fluorescence optimized bathy phytochrome PAiRFP2 derived from wild-type Agp2 in its Pfr state. | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, ... | | Authors: | Sauthof, L, Schmidt, A, Szczepek, M, Scheerer, P. | | Deposit date: | 2018-03-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural snapshot of a bacterial phytochrome in its functional intermediate state.
Nat Commun, 9, 2018
|
|
6G42
 
 | | Crystal structure of mavirus penton protein | | Descriptor: | Minor capsid protein | | Authors: | Born, D, Reuter, L, Meinhart, A, Reinstein, J. | | Deposit date: | 2018-03-26 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Capsid protein structure, self-assembly, and processing reveal morphogenesis of the marine virophage mavirus.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6G6O
 
 | | Crystal structure of the computationally designed Ika8 protein: crystal packing No.1 in P63 | | Descriptor: | GLYCEROL, Ika8 | | Authors: | Noguchi, H, Addy, C, Simoncini, D, Van Meervelt, L, Schiex, T, Zhang, K.Y.J, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2018-04-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Computational design of symmetrical eight-bladed beta-propeller proteins.
IUCrJ, 6, 2019
|
|
6G8A
 
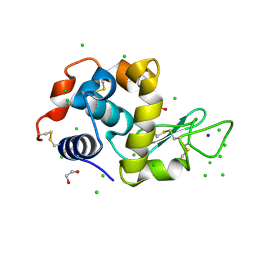 | | Lysozyme solved by Native SAD from a dataset collected in 5 seconds at 1 A wavelength with JUNGFRAU detector | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Leonarski, F, Olieric, V, Vera, L, Redford, S, Wang, M. | | Deposit date: | 2018-04-08 | | Release date: | 2018-08-01 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (1.143 Å) | | Cite: | Fast and accurate data collection for macromolecular crystallography using the JUNGFRAU detector.
Nat. Methods, 15, 2018
|
|
6G8J
 
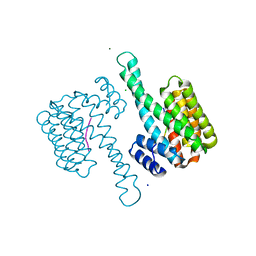 | | 14-3-3sigma in complex with a A130beta3A mutated YAP pS127 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, ACE-ARG-ALA-HIS-SEP-SER-PRO-BAL-SER-LEU-GLN, CHLORIDE ION, ... | | Authors: | Andrei, S.A, Thijssen, V, Brunsveld, L, Ottmann, C, Milroy, L.G. | | Deposit date: | 2018-04-09 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A study on the effect of synthetic alpha-to-beta3-amino acid mutations on the binding of phosphopeptides to 14-3-3 proteins.
Chem.Commun.(Camb.), 55, 2019
|
|
6GA2
 
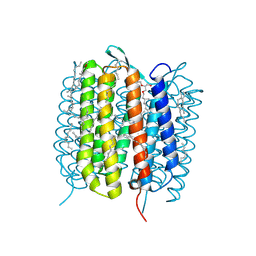 | | Bacteriorhodopsin, dark state, cell 2 | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Barends, T.R.M, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6GA9
 
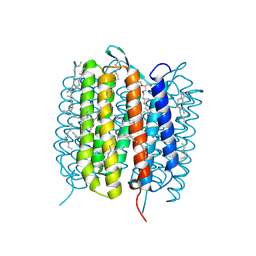 | | BACTERIORHODOPSIN, 390 FS STATE, REAL-SPACE REFINED AGAINST 15% EXTRAPOLATED STRUCTURE FACTORS | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nass Kovacs, G, Colletier, J.-P, Gruenbein, M.L, Stensitzki, T, Batyuk, A, Carbajo, S, Doak, R.B, Ehrenberg, D, Foucar, L, Gasper, R, Gorel, A, Hilpert, M, Kloos, M, Koglin, J, Reinstein, J, Roome, C.M, Schlesinger, R, Seaberg, M, Shoeman, R.L, Stricker, M, Boutet, S, Haacke, S, Heberle, J, Domratcheva, T, Schlichting, I. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional view of ultrafast dynamics in photoexcited bacteriorhodopsin.
Nat Commun, 10, 2019
|
|
6FZ0
 
 | |
6G6X
 
 | | 14-3-3sigma in complex with a P129beta3P mutated YAP pS127 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, SODIUM ION, ... | | Authors: | Andrei, S.A, Thijssen, V, Brunsveld, L, Ottmann, C, Milroy, L.G. | | Deposit date: | 2018-04-03 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | A study on the effect of synthetic alpha-to-beta3-amino acid mutations on the binding of phosphopeptides to 14-3-3 proteins.
Chem.Commun.(Camb.), 55, 2019
|
|
6G1H
 
 | | Amine Dehydrogenase from Petrotoga mobilis; open form | | Descriptor: | 1,2-ETHANEDIOL, Dihydrodipicolinate reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Beloti, L, Frese, A, Mayol, O, Vergne-Vaxelaire, C, Grogan, G. | | Deposit date: | 2018-03-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A family of native amine dehydrogenases for the asymmetric reductive amination of ketones
Nat Catal, 2019
|
|
6G8I
 
 | | 14-3-3sigma in complex with a R124beta3R mutated YAP pS127 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, ALA-HIS-SEP-SER-PRO-ALA-SER-LEU-GLN, CHLORIDE ION, ... | | Authors: | Andrei, S.A, Thijssen, V, Brunsveld, L, Ottmann, C, Milroy, L.G. | | Deposit date: | 2018-04-09 | | Release date: | 2019-04-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A study on the effect of synthetic alpha-to-beta3-amino acid mutations on the binding of phosphopeptides to 14-3-3 proteins.
Chem.Commun.(Camb.), 55, 2019
|
|
6G8Q
 
 | | 14-3-3sigma in complex with a A130beta3A and Q133beta3Q mutated YAP pS127 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Andrei, S.A, Thijssen, V, Brunsveld, L, Ottmann, C, Milroy, L.G. | | Deposit date: | 2018-04-09 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A study on the effect of synthetic alpha-to-beta3-amino acid mutations on the binding of phosphopeptides to 14-3-3 proteins.
Chem.Commun.(Camb.), 55, 2019
|
|
6GDT
 
 | |
6GDU
 
 | | Structure of CutA from Synechococcus elongatus PCC7942 | | Descriptor: | Periplasmic divalent cation tolerance protein | | Authors: | Tremino, L, Rubio, V. | | Deposit date: | 2018-04-24 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Functional and structural characterization of PII-like protein CutA does not support involvement in heavy metal tolerance and hints at a small-molecule carrying/signaling role.
Febs J., 2020
|
|
6FVC
 
 | | Protein environment affects the water-tryptophan binding mode. Molecular dynamics simulations of Engrailed homeodomain mutants | | Descriptor: | Segmentation polarity homeobox protein engrailed | | Authors: | Trosanova, Z, Zachrdla, M, Jansen, S, Srb, P, Zidek, L, Kozelka, J. | | Deposit date: | 2018-03-02 | | Release date: | 2019-04-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Protein environment affects the water-tryptophan binding mode. MD, QM/MM, and NMR studies of engrailed homeodomain mutants.
Phys Chem Chem Phys, 20, 2018
|
|
6GF0
 
 | | Lysozyme structure determined from SFX data using a Sheet-on-Sheet chipless chip | | Descriptor: | Lysozyme C | | Authors: | Doak, R.B, Gorel, A, Foucar, L, Gruenbein, M.L, Hilpert, M, Kloos, M, Nass Kovacs, G, Roome, C, Shoeman, R.L, Stricker, M, Tono, K, You, D, Ueda, K, Sherrel, D, Owen, R, Barends, T.R.M, Schlichting, I. | | Deposit date: | 2018-04-27 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystallography on a chip - without the chip: sheet-on-sheet sandwich.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6GLU
 
 | | Crystal structure of hMTH1 D120N in complex with LW14 in the presence of acetate | | Descriptor: | 1~{H}-imidazo[4,5-b]pyridin-2-amine, 7,8-dihydro-8-oxoguanine triphosphatase | | Authors: | Eberle, S.A, Wiedmer, L, Sledz, P, Caflisch, A. | | Deposit date: | 2018-05-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | hMTH1 D120N in complex with LW14
To Be Published
|
|
6GHX
 
 | |
6GI6
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with a Quinazolinone based ALK2 inhibitor with a 5-methyl core. | | Descriptor: | 1,2-ETHANEDIOL, 5-methyl-6-quinolin-5-yl-3~{H}-quinazolin-4-one, Activin receptor type-1, ... | | Authors: | Williams, E, Hudson, L, Bezerra, G.A, Sorrell, F, Mahajan, P, Kupinska, K, Hoelder, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-05-10 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Novel Quinazolinone Inhibitors of ALK2 Flip between Alternate Binding Modes: Structure-Activity Relationship, Structural Characterization, Kinase Profiling, and Cellular Proof of Concept.
J. Med. Chem., 61, 2018
|
|
6GIH
 
 | | Crystal Structure of CK2alpha with CAM187 bound | | Descriptor: | ACETATE ION, Casein kinase II subunit alpha, [3-chloranyl-5-(1~{H}-indol-4-yl)phenyl]methanamine | | Authors: | Brear, P, Iegre, J, North, A, De Fusco, C, Georgiou, K, Lubin, A, Carro, L, Sore, H, Hyvonen, M, Spring, D. | | Deposit date: | 2018-05-11 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Novel non-ATP competitive small molecules targeting the CK2 alpha / beta interface.
Bioorg. Med. Chem., 26, 2018
|
|
6GLF
 
 | |
6GLL
 
 | | Crystal structure of hMTH1 N33A in complex with LW14 in the presence of acetate | | Descriptor: | 1~{H}-imidazo[4,5-b]pyridin-2-amine, 7,8-dihydro-8-oxoguanine triphosphatase, ACETATE ION, ... | | Authors: | Eberle, S.A, Wiedmer, L, Sledz, P, Caflisch, A. | | Deposit date: | 2018-05-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | hMTH1 N33A in complex with LW14
To Be Published
|
|
6GNV
 
 | | Crystal Structure of Leishmania major N-Myristoyltransferase (NMT) With Bound Myristoyl-CoA and a isopropyl methyl indole aryl sulphonamide ligand | | Descriptor: | GLYCEROL, Glycylpeptide N-tetradecanoyltransferase, TETRADECANOYL-COA, ... | | Authors: | Robinson, D.A, Harrison, J.R, Brand, S, Smith, V.C, Thompson, S, Smith, A, Davies, K, Mok, N.Y, Torrie, L.S, Collie, I, Hallyburton, I, Norval, S, Simeons, F.R.C, Stojanovski, L, Frearson, J.A, Brenk, R, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Molecular Hybridization Approach for the Design of Potent, Highly Selective, and Brain-Penetrant N-Myristoyltransferase Inhibitors.
J. Med. Chem., 61, 2018
|
|
6GPD
 
 | | Crystal structure of the ligand-free form of domain 1 from TmArgBP | | Descriptor: | Amino acid ABC transporter, periplasmic amino acid-binding protein,Amino acid ABC transporter, periplasmic amino acid-binding protein | | Authors: | Smaldone, G, Balasco, N, Ruggiero, A, Berisio, R, Vitagliano, L. | | Deposit date: | 2018-06-05 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Domain communication in Thermotoga maritima Arginine Binding Protein unraveled through protein dissection.
Int. J. Biol. Macromol., 119, 2018
|
|
