1D5Y
 
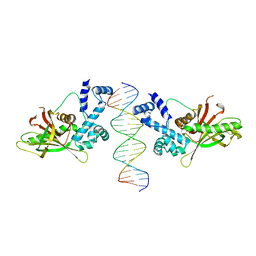 | | CRYSTAL STRUCTURE OF THE E. COLI ROB TRANSCRIPTION FACTOR IN COMPLEX WITH DNA | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*TP*GP*AP*CP*AP*TP*TP*CP*AP*GP*TP*GP*CP*TP*GP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*CP*AP*GP*CP*AP*CP*TP*GP*AP*AP*TP*GP*TP*CP*AP*AP*AP*G)-3'), ROB TRANSCRIPTION FACTOR | | Authors: | Kwon, H.J, Bennik, M.H.J, Demple, B, Ellenberger, T. | | Deposit date: | 1999-10-12 | | Release date: | 2000-04-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA.
Nat.Struct.Biol., 7, 2000
|
|
8GJM
 
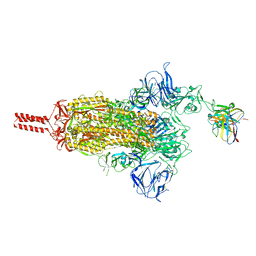 | | 17b10 fab in complex with full-length SARS-CoV-2 Spike G614 trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kwon, H.J, Zhang, J, Kosikova, M, Tang, W.C, Rodriguez, U.O, Peng, H.Q, Meseda, C.A, Pedro, C.L, Schmeisser, F, Lu, J.M, Zhou, B, Davis, C.T, Wentworth, D.E, Chen, W.H, Shriver, M.C, Pasetti, M.F, Weir, J.P, Chen, B, Xie, H. | | Deposit date: | 2023-03-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Distinct in vitro and in vivo neutralization profiles of monoclonal antibodies elicited by the receptor binding domain of the ancestral SARS-CoV-2.
J Med Virol, 95, 2023
|
|
8GJN
 
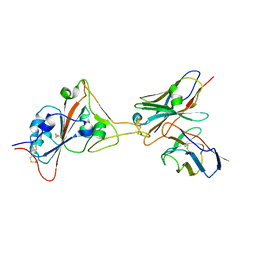 | | 17B10 fab in complex with up-RBD of SARS-CoV-2 Spike G614 trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 17B10 Fab, Light chain of 17B10 Fab, ... | | Authors: | Kwon, H.J, Zhang, J, Kosikova, M, Tang, W.C, Rodriguez, U.O, Peng, H.Q, Meseda, C.A, Pedro, C.L, Schmeisser, F, Lu, J.M, Zhou, B, Davis, C.T, Wentworth, D.E, Chen, W.H, Shriver, M.C, Pasetti, M.F, Weir, J.P, Chen, B, Xie, H. | | Deposit date: | 2023-03-16 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Distinct in vitro and in vivo neutralization profiles of monoclonal antibodies elicited by the receptor binding domain of the ancestral SARS-CoV-2.
J Med Virol, 95, 2023
|
|
3BPS
 
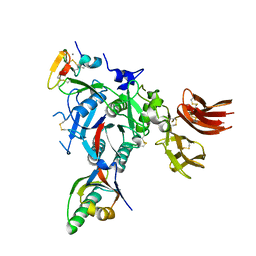 | | PCSK9:EGF-A complex | | Descriptor: | CALCIUM ION, Low-density lipoprotein receptor, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Kwon, H.J. | | Deposit date: | 2007-12-19 | | Release date: | 2008-02-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Molecular basis for LDL receptor recognition by PCSK9.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3QNT
 
 | | NPC1L1 (NTD) Structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Niemann-Pick C1-like protein 1 | | Authors: | Kwon, H.J. | | Deposit date: | 2011-02-09 | | Release date: | 2011-05-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | The Structure of the NPC1L1 N-Terminal Domain in a Closed Conformation.
Plos One, 6, 2011
|
|
1AE9
 
 | |
3GKH
 
 | | NPC1(NTD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Kwon, H.J. | | Deposit date: | 2009-03-10 | | Release date: | 2009-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol.
Cell(Cambridge,Mass.), 137, 2009
|
|
3GCW
 
 | | PCSK9:EGFA(H306Y) | | Descriptor: | CALCIUM ION, Low-density lipoprotein receptor, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Kwon, H.J. | | Deposit date: | 2009-02-22 | | Release date: | 2009-03-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Antagonism of Secreted PCSK9 Increases Low Density Lipoprotein Receptor Expression in HepG2 Cells.
J.Biol.Chem., 284, 2009
|
|
3GKJ
 
 | | NPC1D(NTD):25hydroxycholesterol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 25-HYDROXYCHOLESTEROL, ... | | Authors: | Kwon, H.J. | | Deposit date: | 2009-03-10 | | Release date: | 2009-07-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol.
Cell(Cambridge,Mass.), 137, 2009
|
|
3GKI
 
 | | NPC1(NTD):cholesterol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Kwon, H.J. | | Deposit date: | 2009-03-10 | | Release date: | 2009-07-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol.
Cell(Cambridge,Mass.), 137, 2009
|
|
5TH6
 
 | | Structure determination of a potent, selective antibody inhibitor of human MMP9 (apo MMP9) | | Descriptor: | CALCIUM ION, Matrix metalloproteinase-9,Matrix metalloproteinase-9, ZINC ION | | Authors: | Appleby, T.C, Greenstein, A.E, Kwon, H.J. | | Deposit date: | 2016-09-29 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical characterization and structure determination of a potent, selective antibody inhibitor of human MMP9.
J. Biol. Chem., 292, 2017
|
|
5TH9
 
 | | Structure determination of a potent, selective antibody inhibitor of human MMP9 (GS-5745 bound to MMP-9) | | Descriptor: | CALCIUM ION, COBALT HEXAMMINE(III), GS-5745 Fab heavy chain, ... | | Authors: | Appleby, T.C, Greenstein, A.E, Kwon, H.J. | | Deposit date: | 2016-09-29 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.999 Å) | | Cite: | Biochemical characterization and structure determination of a potent, selective antibody inhibitor of human MMP9.
J. Biol. Chem., 292, 2017
|
|
7TPR
 
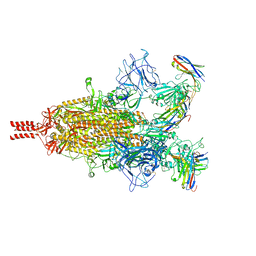 | | Camel nanobodies 7A3 and 8A2 broadly neutralize SARS-CoV-2 variants | | Descriptor: | Nanobody 7A3, Nanobody 8A2, Spike glycoprotein | | Authors: | Butay, K.J, Zhu, J, Dandey, V.P, Hong, J, Kwon, H.J, Chen, C.Z, Duan, Z, Li, D, Ren, H, Liang, T, Martin, N, Esposito, D, Ortega-Rodriguez, U, Xu, M, Xie, H, Ho, M, Cachau, R, Borgnia, M.J. | | Deposit date: | 2022-01-25 | | Release date: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Camel nanobodies broadly neutralize SARS-CoV-2 variants
bioRxiv, 2021
|
|
3ONT
 
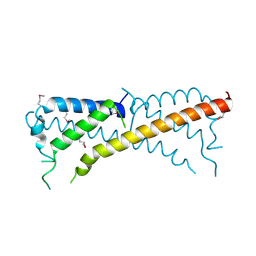 | |
1P7D
 
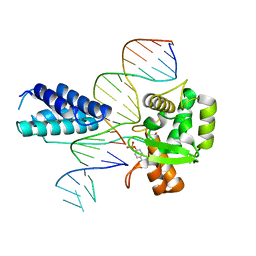 | | Crystal structure of the Lambda Integrase (residues 75-356) bound to DNA | | Descriptor: | 26-MER, 5'-D(*CP*AP*AP*TP*GP*CP*CP*AP*AP*CP*TP*TP*T)-3', Integrase | | Authors: | Aihara, H, Kwon, H.J, Nunes-Duby, S.E, Landy, A, Ellenberger, T. | | Deposit date: | 2003-05-01 | | Release date: | 2003-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A Conformational Switch Controls the DNA Cleavage Activity of Lambda Integrase
Mol.Cell, 12, 2003
|
|
3GCX
 
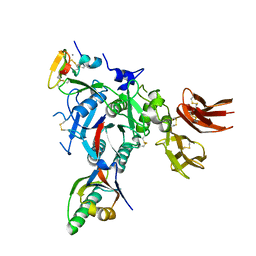 | | PCSK9:EGFA (pH 7.4) | | Descriptor: | CALCIUM ION, Low-density lipoprotein receptor, Proprotein convertase subtilisin/kexin type 9 | | Authors: | McNutt, M.C, Kwon, H.J, Chen, C, Chen, J.R, Horton, J.D, Lagace, T.A. | | Deposit date: | 2009-02-22 | | Release date: | 2009-03-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Antagonism of Secreted PCSK9 Increases Low Density Lipoprotein Receptor Expression in HepG2 Cells.
J.Biol.Chem., 284, 2009
|
|
