2OUB
 
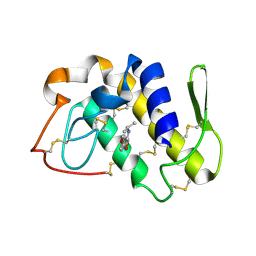 | | Crystal structure of the complex formed between phospholipase A2 and atenolol at 2.75 A resolution | | Descriptor: | 2-(4-(2-HYDROXY-3-(ISOPROPYLAMINO)PROPOXY)PHENYL)ETHANAMIDE, Phospholipase A2 VRV-PL-VIIIa | | Authors: | Kumar, S, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-02-10 | | Release date: | 2007-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of the complex formed between phospholipase A2 and atenolol at 2.75 A resolution
To be Published
|
|
2OTF
 
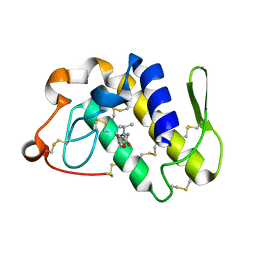 | | Crystal structure of the complex formed between phospholipase A2 and atenolol at 1.95 A resolution | | Descriptor: | 2-(4-(2-HYDROXY-3-(ISOPROPYLAMINO)PROPOXY)PHENYL)ETHANAMIDE, Phospholipase A2 VRV-PL-VIIIa | | Authors: | Kumar, S, Singh, N, Sharma, S, Bhushan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2007-02-08 | | Release date: | 2007-02-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the complex formed between phospholipase A2 and atenolol at 1.95 A resolution
To be Published
|
|
2OYF
 
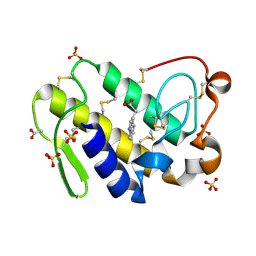 | | Crystal Structure of the complex of phospholipase A2 with indole acetic acid at 1.2 A resolution | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, ACETIC ACID, Phospholipase A2 VRV-PL-VIIIa, ... | | Authors: | Kumar, S, Hariprasad, G, Singh, N, Sharma, S, Kaur, P, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2007-02-22 | | Release date: | 2007-03-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of the complex of phospholipase A2 with indole acetic acid at 1.2 A resolution
To be Published
|
|
2OTH
 
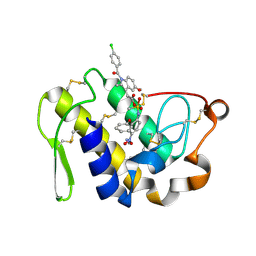 | | Crystal structure of a ternary complex of phospholipase A2 with indomethacin and nimesulide at 2.9 A resolution | | Descriptor: | 4-NITRO-2-PHENOXYMETHANESULFONANILIDE, ACETONITRILE, INDOMETHACIN, ... | | Authors: | Kumar, S, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-02-08 | | Release date: | 2007-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a ternary complex of phospholipase A2 with indomethacin and nimesulide at 2.9 A resolution
To be Published
|
|
3LI6
 
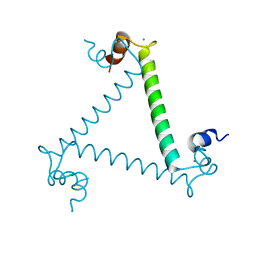 | | Crystal structure and trimer-monomer transition of N-terminal domain of EhCaBP1 from Entamoeba histolytica | | Descriptor: | CALCIUM ION, Calcium-binding protein | | Authors: | Kumar, S, Ahmad, E, Kumar, S, Mansuri, M.S, Khan, R.H, Samudrala, G. | | Deposit date: | 2010-01-24 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystal structure and trimer-monomer transition of N-terminal domain of EhCaBP1 from Entamoeba histolytica
Biophys.J., 98, 2010
|
|
5XOP
 
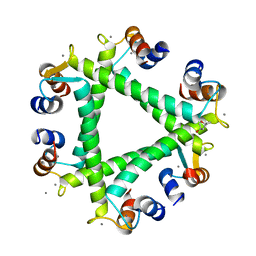 | | Crystal Structure of N-terminal domain EhCaBP1 EF-2 mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Calcium-binding protein 1 (EhCBP1), ... | | Authors: | Kumar, S, Gourinath, S. | | Deposit date: | 2017-05-30 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of calcium binding protein-1 from Entamoeba histolytica: a novel arrangement of EF hand motifs.
Proteins, 68, 2007
|
|
3N94
 
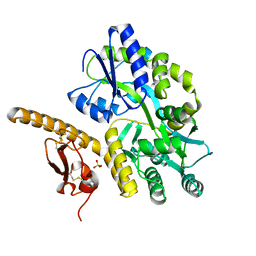 | | Crystal structure of human pituitary adenylate cyclase 1 Receptor-short N-terminal extracellular domain | | Descriptor: | Fusion protein of Maltose-binding periplasmic protein and pituitary adenylate cyclase 1 Receptor-short, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kumar, S, Pioszak, A.A, Swaminathan, K, Xu, H.E. | | Deposit date: | 2010-05-28 | | Release date: | 2011-06-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the PAC1R Extracellular Domain Unifies a Consensus Fold for Hormone Recognition by Class B G-Protein Coupled Receptors.
Plos One, 6, 2011
|
|
3PX1
 
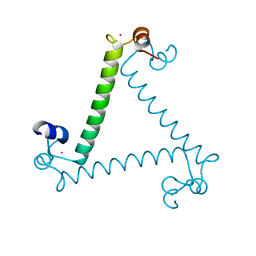 | | Structure of Calcium Binding Protein-1 from Entamoeba histolytica in complex with Strontium | | Descriptor: | Calcium-binding protein, STRONTIUM ION | | Authors: | Kumar, S, Kumar, S, Ahmad, E, Khan, R.H, Gourinath, S. | | Deposit date: | 2010-12-09 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Flexibility and plasticity of EF-hand motifs: Structure of Calcium Binding Protein-1 from Entamoeba histolytica in complex with Pb2+, Ba2+, and Sr2+.
To be Published
|
|
4H3O
 
 | | Crystal structure of a new form of lectin from Allium sativum at 2.17 A resolution | | Descriptor: | CADMIUM ION, Lectin, SODIUM ION | | Authors: | Kumar, S, Yamini, S, Kumar, J, Kaur, P, Singh, T.P, Dey, S. | | Deposit date: | 2012-09-14 | | Release date: | 2012-09-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of a new form of lectin from Allium sativum at 2.17 A resolution
To be Published
|
|
6IDN
 
 | | Crystal structure of ICChI chitinase from ipomoea carnea | | Descriptor: | CALCIUM ION, ICChI, a glycosylated chitinase, ... | | Authors: | Kumar, S, Kumar, A, Patel, A.K. | | Deposit date: | 2018-09-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | TIM barrel fold and glycan moieties in the structure of ICChI, a protein with chitinase and lysozyme activity.
Phytochemistry, 170, 2020
|
|
6KSN
 
 | | Structure of a Zn-bound camelid single domain antibody | | Descriptor: | 1,2-ETHANEDIOL, ICab3, SODIUM ION, ... | | Authors: | Kumar, S, Athreya, A, Penmatsa, A. | | Deposit date: | 2019-08-24 | | Release date: | 2019-11-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Isolation and structural characterization of a Zn2+-bound single-domain antibody against NorC, a putative multidrug efflux transporter in bacteria.
J.Biol.Chem., 295, 2020
|
|
1F9B
 
 | | MELANIN PROTEIN INTERACTION: X-RAY STRUCTURE OF THE COMPLEX OF MARE LACTOFERRIN WITH MELANIN MONOMERS | | Descriptor: | 3H-INDOLE-5,6-DIOL, BICARBONATE ION, FE (III) ION, ... | | Authors: | Kumar, S, Singh, T.P, Sharma, A.K, Singh, N, Raman, G. | | Deposit date: | 2000-07-10 | | Release date: | 2001-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Lactoferrin-melanin interaction and its possible implications in melanin polymerization: crystal structure of the complex formed between mare lactoferrin and melanin monomers at 2.7-A resolution.
Proteins, 45, 2001
|
|
3MU7
 
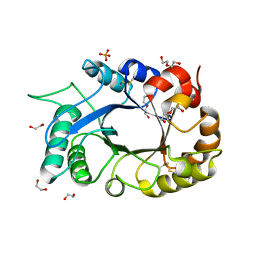 | | Crystal structure of the xylanase and alpha-amylase inhibitor protein (XAIP-II) from scadoxus multiflorus at 1.2 A resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, xylanase and alpha-amylase inhibitor protein | | Authors: | Kumar, S, Singh, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-05-02 | | Release date: | 2010-07-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Modulation of inhibitory activity of xylanase-alpha-amylase inhibitor protein (XAIP): binding studies and crystal structure determination of XAIP-II from Scadoxus multiflorus at 1.2 A resolution.
Bmc Struct.Biol., 10, 2010
|
|
4HZC
 
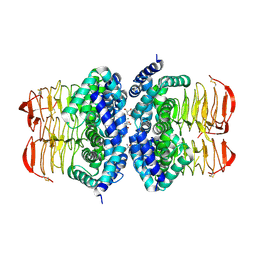 | |
4HZD
 
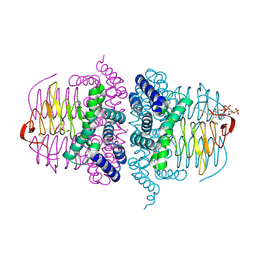 | |
3ULG
 
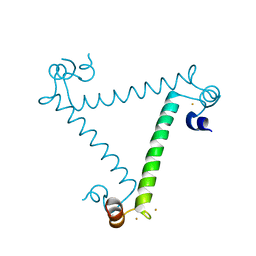 | |
3M7S
 
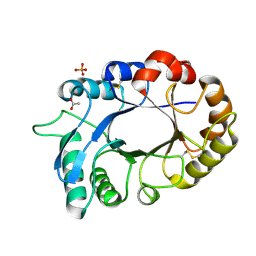 | | Crystal structure of the complex of xylanase GH-11 and alpha amylase inhibitor protein with cellobiose at 2.4 A resolution | | Descriptor: | ACETATE ION, Haementhin, PHOSPHATE ION, ... | | Authors: | Kumar, S, Dube, D, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-03-17 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure determination and inhibition studies of a novel xylanase and alpha-amylase inhibitor protein (XAIP) from Scadoxus multiflorus.
Febs J., 277, 2010
|
|
6U8T
 
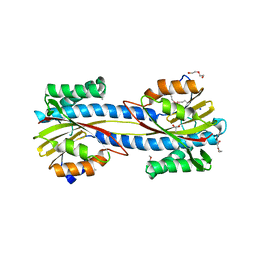 | |
6UUM
 
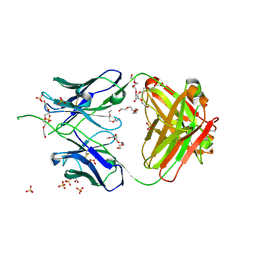 | | Crystal structure of antibody 438-B11 DSS mutant (Cys98A-Cys100aA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, B11 DSS Fab Heavy Chain, ... | | Authors: | Kumar, S, Wilson, I.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A V H 1-69 antibody lineage from an infected Chinese donor potently neutralizes HIV-1 by targeting the V3 glycan supersite.
Sci Adv, 6, 2020
|
|
6UUL
 
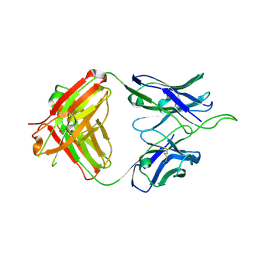 | | Crystal structure of broad and potent HIV-1 neutralizing antibody 438-D5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, D5 Fab Heavy Chain, ... | | Authors: | Kumar, S, Wilson, I.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | A V H 1-69 antibody lineage from an infected Chinese donor potently neutralizes HIV-1 by targeting the V3 glycan supersite.
Sci Adv, 6, 2020
|
|
6UTK
 
 | |
6UUH
 
 | |
6V6W
 
 | |
8DTG
 
 | | Cryo-EM structure of Arabidopsis SPY alternative conformation 1 | | Descriptor: | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | Deposit date: | 2022-07-25 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
8DTI
 
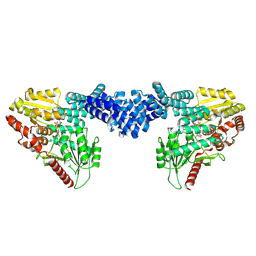 | | Cryo-EM structure of Arabidopsis SPY in complex with GDP-fucose | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-BETA-L-FUCOPYRANOSE, Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | Deposit date: | 2022-07-25 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
