4TOG
 
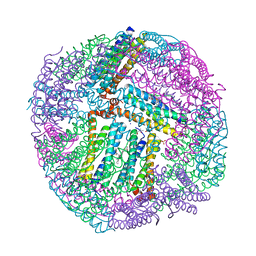 | | 1.80A resolution structure of BfrB (C89S, K96C) crystal form 2 from Pseudomonas aeruginosa | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Bacterioferritin, POTASSIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Yao, H, Kumar, R, Eshelman, K, Rivera, M. | | Deposit date: | 2014-06-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Concerted motions networking pores and distant ferroxidase centers enable bacterioferritin function and iron traffic.
Biochemistry, 54, 2015
|
|
4TOC
 
 | | 2.25A resolution structure of Iron Bound BfrB (Q151L) from Pseudomonas aeruginosa | | Descriptor: | Bacterioferritin, FE (II) ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lovell, S, Battaile, K.P, Yao, H, Kumar, R, Eshelman, K, Rivera, M. | | Deposit date: | 2014-06-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Concerted motions networking pores and distant ferroxidase centers enable bacterioferritin function and iron traffic.
Biochemistry, 54, 2015
|
|
1SQZ
 
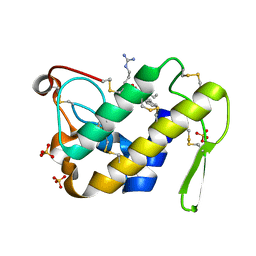 | | Design of specific inhibitors of Phopholipase A2: Crystal structure of the complex formed between Group II Phopholipase A2 and a designed peptide Dehydro-Ile-Ala-Arg-Ser at 1.2A resolution | | Descriptor: | Phospholipase A2, SULFATE ION, synthetic peptide | | Authors: | Singh, N, Prem Kumar, R, Somvanshi, R.K, Bilgrami, S, Ethayathulla, A.S, Sharma, S, Dey, S, Singh, T.P. | | Deposit date: | 2004-03-22 | | Release date: | 2004-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Design of specific inhibitors of Phopholipase A2: Crystal structure of the complex formed between GroupII Phopholipase A2 and a designed peptide Dehydro-Ile-Ala-Arg-Ser at 1.2A resolution
To be Published
|
|
4TOD
 
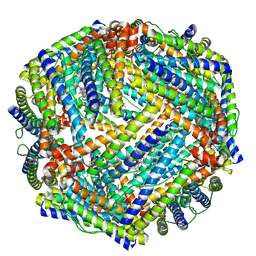 | | 2.05A resolution structure of BfrB (D34F) from Pseudomonas aeruginosa | | Descriptor: | Bacterioferritin, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lovell, S, Battaile, K.P, Yao, H, Kumar, R, Eshelman, K, Rivera, M. | | Deposit date: | 2014-06-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Concerted motions networking pores and distant ferroxidase centers enable bacterioferritin function and iron traffic.
Biochemistry, 54, 2015
|
|
4TOA
 
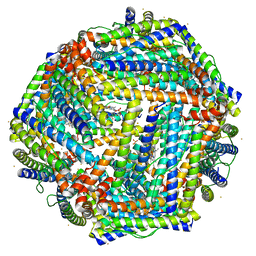 | | 1.95A resolution structure of Iron Bound BfrB (N148L) from Pseudomonas aeruginosa | | Descriptor: | Bacterioferritin, FE (II) ION, POTASSIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Yao, H, Kumar, R, Eshelman, K, Rivera, M. | | Deposit date: | 2014-06-05 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Concerted motions networking pores and distant ferroxidase centers enable bacterioferritin function and iron traffic.
Biochemistry, 54, 2015
|
|
7LXD
 
 | | Structure of yeast DNA Polymerase Zeta (apo) | | Descriptor: | DNA polymerase delta small subunit, DNA polymerase delta subunit 3, DNA polymerase zeta catalytic subunit, ... | | Authors: | Truong, C.D, Craig, T.A, Cui, G, Botuyan, M.V, Serkasevich, R.A, Chan, K.-Y, Mer, G, Chiu, P.-L, Kumar, R. | | Deposit date: | 2021-03-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | Cryo-EM reveals conformational flexibility in apo DNA polymerase zeta.
J.Biol.Chem., 297, 2021
|
|
4E6K
 
 | | 2.0 A resolution structure of Pseudomonas aeruginosa bacterioferritin (BfrB) in complex with bacterioferritin associated ferredoxin (Bfd) | | Descriptor: | Bacterioferritin, FE2/S2 (INORGANIC) CLUSTER, PHOSPHATE ION, ... | | Authors: | Lovell, S, Battaile, K.P, Yao, H, Wang, Y, Kumar, R, Ruvinsky, A, Vasker, I, Rivera, M. | | Deposit date: | 2012-03-15 | | Release date: | 2012-08-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of the BfrB-Bfd Complex Reveals Protein-Protein Interactions Enabling Iron Release from Bacterioferritin.
J.Am.Chem.Soc., 134, 2012
|
|
2E1S
 
 | | Crystal structure of the complex of C-terminal half of bovine lactoferrin and arabinose at 2.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Mir, R, Prem Kumar, R, Singh, N, Sinha, M, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2006-10-27 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the complex of C-terminal half of bovine lactoferrin and arabinose at 2.7 A resolution
To be Published
|
|
2DXR
 
 | | Crystal structure of the complex formed between C-terminal half of bovine lactoferrin and sorbitol at 2.85 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Mir, R, Prem kumar, R, Sinha, M, Singh, N, Sharma, S, Bhushan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2006-08-30 | | Release date: | 2006-09-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of the complex formed between C-terminal half of bovine lactoferrin and sorbitol at 2.85 A resolution
To be Published
|
|
2DYX
 
 | | Structure of the complex of lactoferrin C-lobe with melibiose at 2.0 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Mir, R, Prem kumar, R, Sinha, M, Singh, N, Sharma, S, Kaur, P, Bhushan, A, Singh, T.P. | | Deposit date: | 2006-09-19 | | Release date: | 2006-10-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the complex of lactoferrin C-lobe with melibiose at 2.0 A resolution
To be Published
|
|
2E0S
 
 | | Carbohydrate recognition of C-terminal half of lactoferrin: Crystal structure of the complex of C-lobe with rhamnose at 2.15 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Mir, R, Prem Kumar, R, Singh, N, Sinha, M, Sharma, S, Bhushan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2006-10-11 | | Release date: | 2006-10-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Carbohydrate recognition of C-terminal half of lactoferrin: Crystal structure of the complex of C-lobe with rhamnose at 2.15 A resolution
To be Published
|
|
2HCA
 
 | | Crystal structure of bovine lactoferrin C-lobe liganded with Glucose at 2.8 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Mir, R, Prem Kumar, R, Ethayathulla, A.S, Singh, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2006-06-15 | | Release date: | 2006-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of bovine lactoferrin C-lobe liganded with Glucose at 2.8 A resolution
To be Published
|
|
2H4I
 
 | | Crystal structure of the complex of proteolytically produced C-terminal half of bovine lactoferrin with lactose at 2.55 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Mir, R, Prem kumar, R, Sinha, M, Singh, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2006-05-24 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the complex of proteolytically produced C-terminal half of bovine lactoferrin with lactose at 2.55 A resolution
To be Published
|
|
3ETA
 
 | | Kinase domain of insulin receptor complexed with a pyrrolo pyridine inhibitor | | Descriptor: | 1-(3-{5-[4-(aminomethyl)phenyl]-1H-pyrrolo[2,3-b]pyridin-3-yl}phenyl)-3-(2-phenoxyphenyl)urea, insulin receptor, kinase domain | | Authors: | Patnaik, S, Stevens, K, Gerding, R, Deanda, F, Shotwell, B, Tang, J, Hamajima, T, Nakamura, H, Leesnitzer, A, Hassell, A, Shewchuk, L, Kumar, R, Lei, H, Chamberlain, S. | | Deposit date: | 2008-10-07 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of 3,5-disubstituted-1H-pyrrolo[2,3-b]pyridines as potent inhibitors of the insulin-like growth factor-1 receptor (IGF-1R) tyrosine kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4FAO
 
 | | Specificity and Structure of a high affinity Activin-like 1 (ALK1) signaling complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-2B, Growth/differentiation factor 2, ... | | Authors: | Townson, S.A, Martinez-Hackert, E, Greppi, C, Lowden, P, Sako, D, Liu, J, Ucran, J.A, Liharska, K, Underwood, K.W, Seehra, J, Kumar, R, Grinberg, A.V. | | Deposit date: | 2012-05-22 | | Release date: | 2012-06-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.357 Å) | | Cite: | Specificity and Structure of a High Affinity Activin Receptor-like Kinase 1 (ALK1) Signaling Complex.
J.Biol.Chem., 287, 2012
|
|
4E3R
 
 | | PLP-bound aminotransferase mutant crystal structure from Vibrio fluvialis | | Descriptor: | Pyruvate transaminase, SODIUM ION, SULFATE ION | | Authors: | Midelfort, K.S, Kumar, R, Han, S, Karmilowicz, M.J, McConnell, K, Gehlhaar, D.K, Mistry, A, Chang, J.S, Anderson, M, Vilalobos, A, Minshull, J, Govindarajan, S, Wong, J.W. | | Deposit date: | 2012-03-10 | | Release date: | 2012-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning and characterizing the substrate specificity and activity of Vibrio fluvialis aminotransferase for the synthesis of imagabalin.
Protein Eng.Des.Sel., 26, 2013
|
|
4E3Q
 
 | | PMP-bound form of Aminotransferase crystal structure from Vibrio fluvialis | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, BENZAMIDINE, Pyruvate transaminase, ... | | Authors: | Midelfort, K.S, Kumar, R, Han, S, Karmilowicz, M.J, McConnell, K, Gehlhaar, D.K, Mistry, A, Chang, J.S, Anderson, M, Vilalobos, A, Minshull, J, Govindarajan, S, Wong, J.W. | | Deposit date: | 2012-03-10 | | Release date: | 2012-10-10 | | Last modified: | 2013-01-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning and characterizing the substrate specificity and activity of Vibrio fluvialis aminotransferase for the synthesis of imagabalin.
Protein Eng.Des.Sel., 26, 2013
|
|
3RUV
 
 | | Crystal structure of Cpn-rls in complex with ATP analogue from Methanococcus maripaludis | | Descriptor: | Chaperonin, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Pereira, J.H, Ralston, C.Y, Douglas, N.R, Kumar, R, McAndrew, R.P, Knee, K.M, King, J.A, Frydman, J, Adams, P.D. | | Deposit date: | 2011-05-05 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Mechanism of nucleotide sensing in group II chaperonins.
Embo J., 31, 2012
|
|
3RUQ
 
 | | Crystal structure of Cpn-WT in complex with ADP from Methanococcus maripaludis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin, MAGNESIUM ION | | Authors: | Pereira, J.H, Ralston, C.Y, Douglas, N.R, Kumar, R, McAndrew, R.P, Knee, K.M, Frydman, J, Adams, P.D. | | Deposit date: | 2011-05-05 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Mechanism of nucleotide sensing in group II chaperonins.
Embo J., 31, 2012
|
|
3RUS
 
 | | Crystal structure of Cpn-rls in complex with ADP from Methanococcus maripaludis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin, MAGNESIUM ION, ... | | Authors: | Pereira, J.H, Ralston, C.Y, Douglas, N.R, Kumar, R, McAndrew, R.P, Knee, K.M, King, J.A, Frydman, J, Adams, P.D. | | Deposit date: | 2011-05-05 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.338 Å) | | Cite: | Mechanism of nucleotide sensing in group II chaperonins.
Embo J., 31, 2012
|
|
3RUW
 
 | | Crystal structure of Cpn-rls in complex with ADP-AlFx from Methanococcus maripaludis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Chaperonin, ... | | Authors: | Pereira, J.H, Ralston, C.Y, Douglas, N.R, Kumar, R, McAndrew, R.P, Knee, K.M, King, J.A, Frydman, J, Adams, P.D. | | Deposit date: | 2011-05-05 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Mechanism of nucleotide sensing in group II chaperonins.
Embo J., 31, 2012
|
|
1ZWP
 
 | | The atomic resolution Crystal structure of the Phospholipase A2 (PLA2) complex with Nimesulide reveals its weaker binding to PLA2 | | Descriptor: | 4-NITRO-2-PHENOXYMETHANESULFONANILIDE, METHANOL, Phospholipase A2 VRV-PL-VIIIa, ... | | Authors: | Prem Kumar, R, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2005-06-04 | | Release date: | 2005-07-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The atomic resolution Crystal structure of the Phospholipase A2 (PLA2) complex with Nimesulide reveals its weaker binding to PLA2
To be Published
|
|
6WKD
 
 | | Crystal structure of pentalenene synthase complexed with 12,13-difluorofarnesyl diphosphate | | Descriptor: | (2E,6E)-12-fluoro-11-(fluoromethyl)-3,7-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Prem Kumar, R, Matos, J.O, Oprian, D.D. | | Deposit date: | 2020-04-16 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism Underlying Anti-Markovnikov Addition in the Reaction of Pentalenene Synthase.
Biochemistry, 59, 2020
|
|
6WKC
 
 | |
6WKF
 
 | | Crystal structure of pentalenene synthase mutant F76Y complexed with 12,13-difluorofarnesyl diphosphate | | Descriptor: | (2E,6E)-12-fluoro-11-(fluoromethyl)-3,7-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, Pentalenene synthase | | Authors: | Prem Kumar, R, Matos, J.O, Oprian, D.D. | | Deposit date: | 2020-04-16 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism Underlying Anti-Markovnikov Addition in the Reaction of Pentalenene Synthase.
Biochemistry, 59, 2020
|
|
