5R7Z
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z1220452176 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{N}-[2-(5-fluoranyl-1~{H}-indol-3-yl)ethyl]ethanamide | | Authors: | Fearon, D, Powell, A.J, Douangamath, A, Owen, C.D, Wild, C, Krojer, T, Lukacik, P, Strain-Damerell, C.M, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-03 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5REF
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z24758179 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, methyl 3-(methylsulfonylamino)benzoate | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RET
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102269 | | Descriptor: | 1-{4-[(3-chlorophenyl)methyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RFC
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z979145504 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, methyl (2-methyl-4-phenyl-1,3-thiazol-5-yl)carbamate | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RFO
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102972 | | Descriptor: | 1-[4-(piperidine-1-carbonyl)piperidin-1-yl]ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RG3
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with NCL-00025412 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N~2~-acetyl-N~1~-prop-2-en-1-yl-L-aspartamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-04-15 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RL9
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z1703168683 | | Descriptor: | 1-(3-fluoro-4-methylphenyl)methanesulfonamide, Helicase, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.788 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RLS
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z59181945 | | Descriptor: | Helicase, N-hydroxyquinoline-2-carboxamide, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.278 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
3LXJ
 
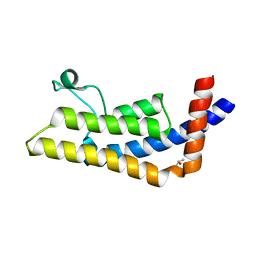 | | Crystal Structure of the Bromodomain of Human AAA domain containing 2B (ATAD2B) | | Descriptor: | ATPase family AAA domain-containing protein 2B, ISOPROPYL ALCOHOL | | Authors: | Filippakopoulos, P, Keates, T, Picaud, S, Fedorov, O, Krojer, T, Vollmar, M, Muniz, J, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-02-25 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
5LF9
 
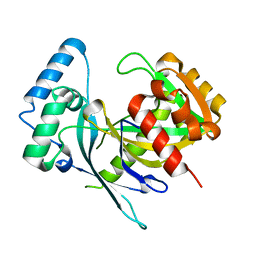 | | Crystal structure of human NUDT22 | | Descriptor: | Nucleoside diphosphate-linked moiety X motif 22 | | Authors: | Tallant, C, Siejka, P, Mathea, S, Shrestha, L, Krojer, T, Srikannathasan, V, Elkins, J.M, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-30 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of human NUDT22
To Be Published
|
|
3MQM
 
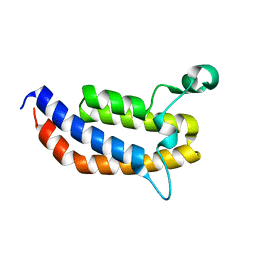 | | Crystal Structure of the Bromodomain of human ASH1L | | Descriptor: | Probable histone-lysine N-methyltransferase ASH1L | | Authors: | Filippakopoulos, P, Picaud, S, Keates, T, Felletar, I, Vollmar, M, Chaikuad, A, Krojer, T, Canning, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-28 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
6FCX
 
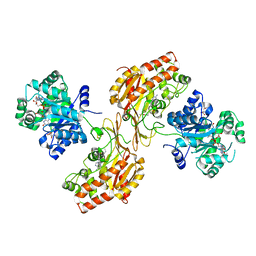 | | Structure of human 5,10-methylenetetrahydrofolate reductase (MTHFR) | | Descriptor: | CITRIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Methylenetetrahydrofolate reductase, ... | | Authors: | Kopec, J, Bezerra, G.A, Oberholzer, A.E, Rembeza, E, Sorrell, F.J, Chalk, R, Borkowska, O, Ellis, K, Kupinska, K, Krojer, T, Burgess-Brown, N, Von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Froese, D.S, Baumgartner, M, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-21 | | Release date: | 2018-05-16 | | Last modified: | 2018-07-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the regulation of human 5,10-methylenetetrahydrofolate reductase by phosphorylation and S-adenosylmethionine inhibition.
Nat Commun, 9, 2018
|
|
5MKV
 
 | | Crystal Structure of Human Dihydropyrimidinease-like 2 (DPYSL2A)/Collapsin Response Mediator Protein (CRMP2) residues 13-516 | | Descriptor: | 1,2-ETHANEDIOL, Dihydropyrimidinase-related protein 2 | | Authors: | Sethi, R, Zheng, Y, Krojer, T, Velupillai, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Ahmed, A.A, von Delft, F. | | Deposit date: | 2016-12-05 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tuning microtubule dynamics to enhance cancer therapy by modulating FER-mediated CRMP2 phosphorylation.
Nat Commun, 9, 2018
|
|
5MR8
 
 | | Crystal structure of TRIM33 PHD-Bromodomain isoform B in complex with H3K9ac histone peptide | | Descriptor: | E3 ubiquitin-protein ligase TRIM33, Histone H3, ZINC ION | | Authors: | Tallant, C, Savitsky, P, Fedorov, O, Nunez-Alonso, G, Siejka, P, Krojer, T, Williams, E, Srikannathasan, V, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Muller, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of TRIM33 PHD-Bromodomain isoform B in complex with H3K9ac histone peptide
To Be Published
|
|
6G0S
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated SIRT7 peptide (K272ac/K275ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, NAD-dependent protein deacetylase sirtuin-7 | | Authors: | Filippakopoulos, P, Picaud, S, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2018-03-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
3S95
 
 | | Crystal structure of the human LIMK1 kinase domain in complex with staurosporine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Beltrami, A, Chaikuad, A, Daga, N, Elkins, J.M, Mahajan, P, Savitsky, P, Vollmar, M, Krojer, T, Muniz, J.R.C, Fedorov, O, Allerston, C.K, Yue, W.W, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-05-31 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the human LIMK1 kinase domain in complex with staurosporine
To be Published
|
|
5AMF
 
 | | Crystal structure of the bromodomain of human surface epitope engineered BRD1A in complex with 3D Consortium fragment Ethyl 4,5,6,7- tetrahydro-1H-indazole-5-carboxylate (SGC - Diamond I04-1 fragment screening) | | Descriptor: | BROMODOMAIN-CONTAINING PROTEIN 1, ETHYL (5R)-4,5,6,7-TETRAHYDRO-1H-INDAZOLE-5-CARBOXYLATE, SODIUM ION | | Authors: | Pearce, N.M, Fairhead, M, Strain-Damerell, C, Talon, R, Wright, N, Ng, J.T, Bradley, A, Cox, O, Bowkett, D, Collins, P, Brandao-Neto, J, Douangamath, A, Krojer, T, Burgess-Brown, N, Brennan, P, Arrowsmith, C.H, Edwards, E, Bountra, C, von Delft, F. | | Deposit date: | 2015-03-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the Bromodomain of Human Surface Epitope Engineered Brd1A in Complex with 3D Consortium Fragment Ethyl 4,5,6,7-Tetrahydro-1H-Indazole-5-Carboxylate
To be Published
|
|
5AME
 
 | | Crystal structure of the bromodomain of human surface epitope engineered BRD1A in complex with 3D Consortium fragment 4-acetyl- piperazin-2-one (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 4-acetyl-piperazin-2-one, BROMODOMAIN-CONTAINING PROTEIN 1, SODIUM ION | | Authors: | Pearce, N.M, Fairhead, M, Strain-Damerell, C, Talon, R, Wright, N, Ng, J.T, Bradley, A, Cox, O, Bowkett, D, Collins, P, Brandao-Neto, J, Douangamath, A, Krojer, T, Burgess-Brown, N, Brennan, P, Arrowsmith, C.H, Edwards, E, Bountra, C, von Delft, F. | | Deposit date: | 2015-03-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.578 Å) | | Cite: | Crystal Structure of the Bromodomain of Human Surface Epitope Engineered Brd1A in Complex with 3D Consortium Fragment 4-Acetyl-Piperazin-2-One
To be Published
|
|
6G0P
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated E2F1 peptide (K117ac/K120ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Transcription factor E2F1 | | Authors: | Filippakopoulos, P, Picaud, S, Krojer, T, Sorrell, F, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2018-03-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
5RH4
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with Z1530425063 (Mpro-x2659) | | Descriptor: | (2R)-2-(6-chloro-9H-carbazol-2-yl)propanoic acid, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-05-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen
To Be Published
|
|
5RJP
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with NCL-00024672 | | Descriptor: | 4-bromo-1-(2-hydroxyethyl)pyridin-2(1H)-one, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.242 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RK3
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z1501469697 | | Descriptor: | 3-amino-1,6-dimethylpyridin-2(1H)-one, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RH7
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with Z4439011584 (Mpro-x2705) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(5-tert-butyl-1H-pyrazol-3-yl)-N-[(1R)-2-[(2-ethyl-6-methylphenyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-05-15 | | Release date: | 2020-05-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen
To Be Published
|
|
5RKN
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z373768900 | | Descriptor: | N-(1-ethyl-1H-pyrazol-4-yl)cyclobutanecarboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RJW
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z256709556 | | Descriptor: | 3-methylthiophene-2-carboxylic acid, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.514 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
