4PY6
 
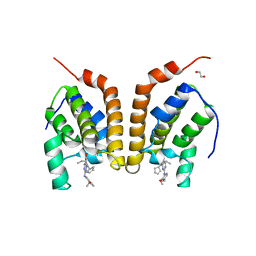 | | Crystal Structure of bromodomain of PFA0510w from Plasmodium Falciparum | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Bromodomain protein, ... | | Authors: | Fonseca, M, Tallant, C, Hutchinson, A, Savitsky, P, Krojer, T, Filippakopoulos, P, Loppnau, P, Brennan, P.E, von Delft, F, Dong, A, Josling, G.A, Duffy, M.F, Arrowsmith, C.H, Bountra, C, Hui, R, Knapp, S, Wernimont, A.K, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-03-26 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of bromodomain of PFA0510w from Plasmodium Falciparum
To be Published
|
|
7PSP
 
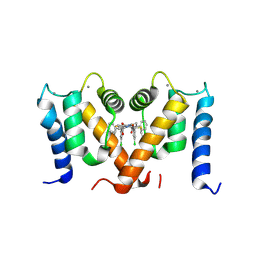 | | Crystal structure of S100A4 labeled with NU000846b. | | Descriptor: | (2R,4R)-1-(2-chloranylethanoyl)-N-(3-chlorophenyl)-4-phenyl-pyrrolidine-2-carboxamide, CALCIUM ION, Protein S100-A4 | | Authors: | Giroud, C, Szommer, T, Coxon, C, Monteiro, O, Christott, T, Bennett, J, Aitmakhanova, K, Raux, B, Newman, J, Elkins, J, Arruda Bezerra, G, Krojer, T, Koekemoer, L, Von Delft, F, Bountr, C, Brennan, P, Fedorov, O. | | Deposit date: | 2021-09-23 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of S100A4 labeled with NU000846b.
To Be Published
|
|
6Q94
 
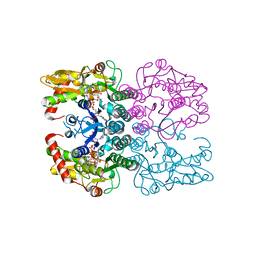 | | Crystal structure of human GDP-D-mannose 4,6-dehydratase (S156D) in complex with GDP-Man | | Descriptor: | 1,2-ETHANEDIOL, GDP-mannose 4,6 dehydratase, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, ... | | Authors: | Pfeiffer, M, Krojer, T, Johansson, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-17 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Parsimonious Mechanism of Sugar Dehydration by Human GDP-Mannose-4,6-dehydratase.
Acs Catalysis, 9, 2019
|
|
4UUC
 
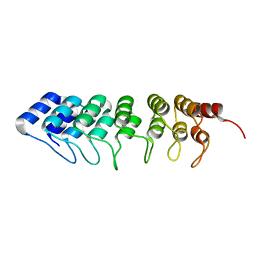 | | Crystal structure of human ASB11 ankyrin repeat domain | | Descriptor: | ANKYRIN REPEAT AND SOCS BOX PROTEIN 11 | | Authors: | Pinkas, D.M, Sanvitale, C, Kragh Nielsen, T, Guo, K, Sorrell, F, Berridge, G, Ayinampudi, V, Wang, D, Newman, J.A, Tallant, C, Chaikuad, A, Canning, P, Kopec, J, Krojer, T, Vollmar, M, Allerston, C.K, Chalk, R, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Bullock, A. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Human Asb11 Ankyrin Repeat Domain
To be Published
|
|
6QY7
 
 | | Human CSNK2A1 bound to ERB-041 | | Descriptor: | 2-(3-FLUORO-4-HYDROXYPHENYL)-7-VINYL-1,3-BENZOXAZOL-5-OL, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Abdul Azeez, K.R, Sorrell, F.J, Krojer, T, Bountra, C, Edwards, A.M, Arrowsmith, C, Knapp, S, Elkins, J.M. | | Deposit date: | 2019-03-08 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CSNK2A1 bound to ERB-041
To Be Published
|
|
6QY9
 
 | | Human CSNK2A2 bound to a Pyrrolo[2,3-d]pyrimidinyl inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 3-[3-[2-[(3,4,5-trimethoxyphenyl)amino]pyrrolo[2,3-d]pyrimidin-7-yl]phenyl]propanenitrile, CHLORIDE ION, ... | | Authors: | Abdul Azeez, K.R, Sorrell, F.J, Krojer, T, Bountra, C, Edwards, A.M, Arrowsmith, C, Knapp, S, Elkins, J.M. | | Deposit date: | 2019-03-08 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Human CSNK2A2 bound to a Pyrrolo[2,3-d]pyrimidinyl inhibitor
To Be Published
|
|
6QY8
 
 | | Human CSNK2A2 bound to ERB-041 | | Descriptor: | 2-(3-FLUORO-4-HYDROXYPHENYL)-7-VINYL-1,3-BENZOXAZOL-5-OL, Casein kinase II subunit alpha' | | Authors: | Abdul Azeez, K.R, Sorrell, F.J, Krojer, T, Bountra, C, Edwards, A.M, Arrowsmith, C, Knapp, S, Elkins, J.M. | | Deposit date: | 2019-03-08 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CSNK2A1 bound to ERB-041
To Be Published
|
|
4MHV
 
 | | Crystal structure of the PNT domain of human ETS2 | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Newman, J.A, Cooper, C.D.O, Krojer, T, Shrestha, L, Burgess-brown, N, Arrowsmith, C.H, Bountra, C, Edwards, A, Gileadi, O. | | Deposit date: | 2013-08-30 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of the PNT domain of human ETS2
To be Published
|
|
6F58
 
 | | Crystal structure of human Brachyury (T) in complex with DNA | | Descriptor: | Brachyury protein, DNA (5'-D(*AP*AP*TP*TP*TP*CP*AP*CP*AP*CP*CP*TP*AP*GP*GP*TP*GP*TP*GP*AP*AP*AP*TP*T)-3'), SODIUM ION | | Authors: | Newman, J.A, Gavard, A.E, Krojer, T, Shrestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-12-01 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | Crystal structure of human Brachyury (T) in complex with DNA
To Be Published
|
|
2WZK
 
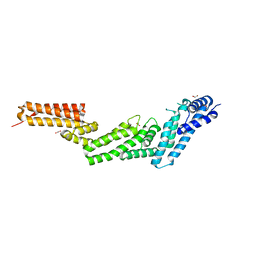 | | Structure of the Cul5 N-terminal domain at 2.05A resolution. | | Descriptor: | 1,2-ETHANEDIOL, CULLIN-5 | | Authors: | Muniz, J.R.C, Ayinampudi, V, Zhang, Y, Babon, J.J, Chaikuad, A, Krojer, T, Pike, A.C.W, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Bullock, A.N. | | Deposit date: | 2009-11-30 | | Release date: | 2009-12-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular Architecture of the Ankyrin Socs Box Family of Cul5-Dependent E3 Ubiquitin Ligases
J.Mol.Biol., 425, 2013
|
|
4MX2
 
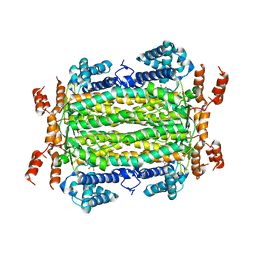 | | Crystal Structure of adenylosuccinate lyase from Leishmania donovani | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenylosuccinate lyase, GLYCEROL, ... | | Authors: | Wernimont, A.K, Loppnau, P, Dong, A, Krojer, T, Bradley, A, Bushell, S, von Delft, F, Robinson, D, Gilbert, I, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Mottaghi, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-09-25 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of adenylosuccinate lyase from Leishmania donovani
To be Published
|
|
2XIO
 
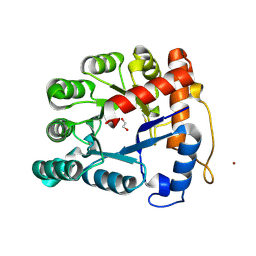 | | Structure of putative deoxyribonuclease TATDN1 isoform a | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, PUTATIVE DEOXYRIBONUCLEASE TATDN1, ... | | Authors: | Muniz, J.R.C, Cocking, R, Puranik, S, Krojer, T, Raynor, J, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Yue, W.W, Kavanagh, K.L, Oppermann, U. | | Deposit date: | 2010-06-30 | | Release date: | 2010-07-14 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structure of Putative Deoxyribonuclease Tatdn1 Isoform A
To be Published
|
|
2XST
 
 | | Crystal Structure of the Human Lipocalin 15 | | Descriptor: | 1,2-ETHANEDIOL, LIPOCALIN 15 | | Authors: | Muniz, J.R.C, Gileadi, C, Yue, W.W, Krojer, T, Ugochukwu, E, Phillips, C, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Kavanagh, K.L, Oppermann, U. | | Deposit date: | 2010-09-30 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structure of the Human Lipocalin 15
To be Published
|
|
2Y1H
 
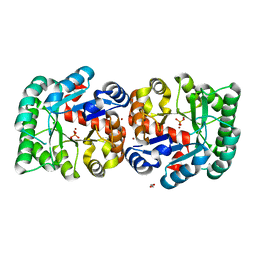 | | Crystal structure of the human TatD-domain protein 3 (TATDN3) | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, PHOSPHATE ION, ... | | Authors: | Muniz, J.R.C, Puranik, S, Krojer, T, Yue, W.W, Ugochukwu, E, Filippakopoulos, P, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Kavanagh, K.L, Oppermann, U. | | Deposit date: | 2010-12-08 | | Release date: | 2010-12-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Human Tatd-Domain Protein 3 (Tatdn3)
To be Published
|
|
2Y7J
 
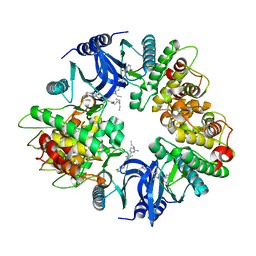 | | Structure of human phosphorylase kinase, gamma 2 | | Descriptor: | N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide, PHOSPHORYLASE B KINASE GAMMA CATALYTIC CHAIN, TESTIS/LIVER ISOFORM | | Authors: | Muniz, J.R.C, Shrestha, A, Savitsky, P, Wang, J, Rellos, P, Fedorov, O, Burgess-Brown, N, Brenner, B, Berridge, G, Elkins, J.M, Krojer, T, Vollmar, M, Che, K.H, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S. | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Human Phosphorylase Kinase, Gamma 2
To be Published
|
|
6F59
 
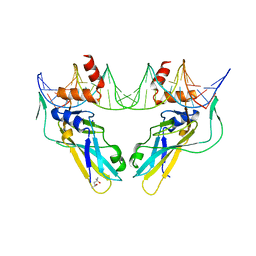 | | Crystal structure of human Brachyury (T) G177D variant in complex with DNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Brachyury protein, DNA (26-MER), ... | | Authors: | Newman, J.A, Gavard, A.E, Krojer, T, Shrestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-12-01 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Crystal structure of human Brachyury (T) G177D variant in complex with DNA
To Be Published
|
|
2YL2
 
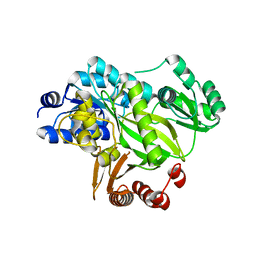 | | Crystal structure of human acetyl-CoA carboxylase 1, biotin carboxylase (BC) domain | | Descriptor: | ACETYL-COA CARBOXYLASE 1 | | Authors: | Muniz, J.R.C, Froese, D.S, Krysztofinska, E, Vollmar, M, Beltrami, A, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Yue, W.W, Oppermann, U. | | Deposit date: | 2011-05-31 | | Release date: | 2011-06-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Human Acetyl-Coa Carboxylase 1, Biotin Carboxylase (Bc) Domain
To be Published
|
|
6TTK
 
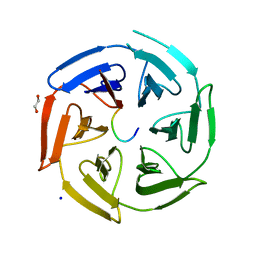 | | Crystal structure of the kelch domain of human KLHL12 in complex with DVL1 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DVL1, ... | | Authors: | Chen, Z, Williams, E, Pike, A.C.W, Strain-Damerell, C, Wang, D, Chalk, R, Burgess-Brown, N, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-12-27 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | Identification of a PGXPP degron motif in dishevelled and structural basis for its binding to the E3 ligase KLHL12.
Open Biology, 10, 2020
|
|
5S20
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with PB1827975385 | | Descriptor: | (5R)-5-amino-5,6,7,8-tetrahydronaphthalen-1-ol, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.037 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S2K
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z445856640 | | Descriptor: | N-[(3R)-1,1-dioxo-1lambda~6~-thiolan-3-yl]-N-methyl-N'-propan-2-ylurea, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.097 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S30
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z65532537 | | Descriptor: | (2R)-2-(2-fluorophenoxy)propanoic acid, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S3K
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z219104216 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 6-(ethylamino)pyridine-3-carbonitrile, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S46
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z57131035 | | Descriptor: | Non-structural protein 3, imidazolidine-2,4-dione | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.191 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S4K
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with FMOOA000509a | | Descriptor: | (2S,5R,6R)-7-methyl-2,3,4,5,6,7-hexahydro-1H-2,6-methanoazocino[5,4-b]indol-5-ol, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.076 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S1G
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with EN300-108952 | | Descriptor: | (4-methylpyridin-3-yl)methanol, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
