2XRQ
 
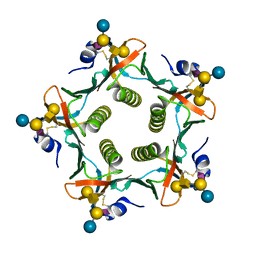 | | Crystal structures exploring the origins of the broader specificity of escherichia coli heat-labile enterotoxin compared to cholera toxin | | Descriptor: | HEAT-LABILE ENTEROTOXIN B CHAIN, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Holmner, A, Mackenzie, A, Okvist, M, Jansson, L, Lebens, M, Teneberg, S, Krengel, U. | | Deposit date: | 2010-09-19 | | Release date: | 2010-12-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures Exploring the Origins of the Broader Specificity of Escherichia Coli Heat-Labile Enterotoxin Compared to Cholera Toxin
J.Mol.Biol., 406, 2011
|
|
2XKN
 
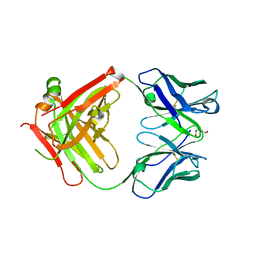 | | Crystal structure of the Fab fragment of the anti-EGFR antibody 7A7 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ANTI-EGFR ANTIBODY 7A7 | | Authors: | Talavera, A, Mackenzie, J, Friemann, R, Krengel, U. | | Deposit date: | 2010-07-09 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the Fab Fragment of the Anti-Murine Egfr Antibody 7A7 and Exploration of its Receptor Binding Site.
Mol.Immunol., 48, 2011
|
|
1X13
 
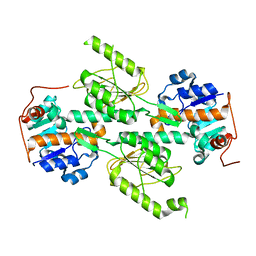 | | Crystal structure of E. coli transhydrogenase domain I | | Descriptor: | NAD(P) transhydrogenase subunit alpha | | Authors: | Johansson, T, Oswald, C, Pedersen, A, Tornroth, S, Okvist, M, Karlsson, B.G, Rydstrom, J, Krengel, U. | | Deposit date: | 2005-03-31 | | Release date: | 2005-09-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray Structure of Domain I of the Proton-pumping Membrane Protein Transhydrogenase from Escherichia coli
J.Mol.Biol., 352, 2005
|
|
1X15
 
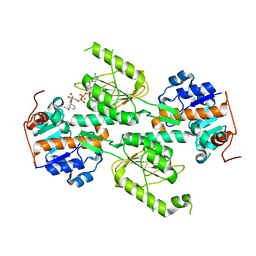 | | Crystal structure of E. coli transhydrogenase domain I with bound NADH | | Descriptor: | NAD(P) transhydrogenase subunit alpha, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Johansson, T, Oswald, C, Pedersen, A, Tornroth, S, Okvist, M, Karlsson, B.G, Rydstrom, J, Krengel, U. | | Deposit date: | 2005-03-31 | | Release date: | 2005-09-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | X-ray Structure of Domain I of the Proton-pumping Membrane Protein Transhydrogenase from Escherichia coli
J.Mol.Biol., 352, 2005
|
|
2FP2
 
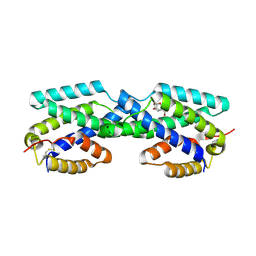 | | Secreted Chorismate Mutase from Mycobacterium tuberculosis | | Descriptor: | 8-HYDROXY-2-OXA-BICYCLO[3.3.1]NON-6-ENE-3,5-DICARBOXYLIC ACID, Chorismate mutase | | Authors: | Okvist, M, Dey, R, Sasso, S, Grahn, E, Kast, P, Krengel, U. | | Deposit date: | 2006-01-15 | | Release date: | 2006-03-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | 1.6A Crystal Structure of the Secreted Chorismate Mutase from Mycobacterium tuberculosis: Novel Fold Topology Revealed
J.Mol.Biol., 357, 2006
|
|
2IHO
 
 | | Crystal structure of MOA, a lectin from the mushroom Marasmius oreades in complex with the trisaccharide Gal(1,3)Gal(1,4)GlcNAc | | Descriptor: | Lectin, beta-D-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Grahn, E, Askarieh, G, Holmner, A, Tateno, H, Winter, H.C, Goldstein, I.J, Krengel, U. | | Deposit date: | 2006-09-27 | | Release date: | 2007-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structure of the marasmius oreades mushroom lectin in complex with a xenotransplantation epitope.
J.Mol.Biol., 369, 2007
|
|
2FP1
 
 | | Secreted Chorismate Mutase from Mycobacterium tuberculosis | | Descriptor: | Chorismate mutase, LEAD (II) ION | | Authors: | Okvist, M, Dey, R, Sasso, S, Grahn, E, Kast, P, Krengel, U. | | Deposit date: | 2006-01-15 | | Release date: | 2006-03-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.6A Crystal Structure of the Secreted Chorismate Mutase from Mycobacterium tuberculosis: Novel Fold Topology Revealed
J.Mol.Biol., 357, 2006
|
|
1X14
 
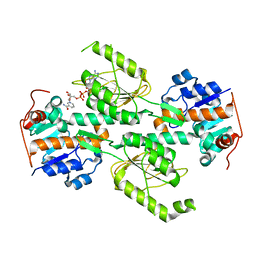 | | Crystal structure of E. coli transhydrogenase domain I with bound NAD | | Descriptor: | NAD(P) transhydrogenase subunit alpha, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Johansson, T, Oswald, C, Pedersen, A, Tornroth, S, Okvist, M, Karlsson, B.G, Rydstrom, J, Krengel, U. | | Deposit date: | 2005-03-31 | | Release date: | 2005-09-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | X-ray Structure of Domain I of the Proton-pumping Membrane Protein Transhydrogenase from Escherichia coli
J.Mol.Biol., 352, 2005
|
|
5HUC
 
 | | DAHP synthase from Corynebacterium glutamicum | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-Deoxy-D-arabino-heptulosonate 7-phosphate (DAHP) synthase, MANGANESE (II) ION, ... | | Authors: | Burschowsky, D, Heim, J.B, Thorbjoernsrud, H.V, Krengel, U. | | Deposit date: | 2016-01-27 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Inter-Enzyme Allosteric Regulation of Chorismate Mutase in Corynebacterium glutamicum: Structural Basis of Feedback Activation by Trp.
Biochemistry, 57, 2018
|
|
5HUB
 
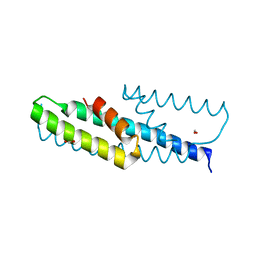 | | High-resolution structure of chorismate mutase from Corynebacterium glutamicum | | Descriptor: | Chorismate mutase, FORMIC ACID | | Authors: | Burschowsky, D, Heim, J.B, Thorbjoernsrud, H.V, Krengel, U. | | Deposit date: | 2016-01-27 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Inter-Enzyme Allosteric Regulation of Chorismate Mutase in Corynebacterium glutamicum: Structural Basis of Feedback Activation by Trp.
Biochemistry, 57, 2018
|
|
5HUE
 
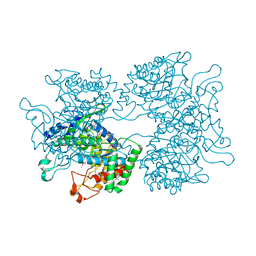 | | DAHP synthase from Corynebacterium glutamicum in complex with tryptophan | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-Deoxy-D-arabino-heptulosonate 7-phosphate (DAHP) synthase, ... | | Authors: | Burschowsky, D, Heim, J.B, Thorbjoernsrud, H.V, Krengel, U. | | Deposit date: | 2016-01-27 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Inter-Enzyme Allosteric Regulation of Chorismate Mutase in Corynebacterium glutamicum: Structural Basis of Feedback Activation by Trp.
Biochemistry, 57, 2018
|
|
5LZI
 
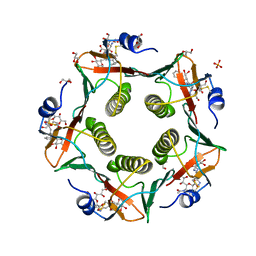 | | Porcine heat-labile enterotoxin R13H in complex with inhibitor MM146 | | Descriptor: | (2~{R},4~{S},5~{R},6~{R})-5-acetamido-2-[4-[(2~{R})-3-[2-[(2~{S},3~{R},4~{R},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]ethylamino]-3-oxidanylidene-2-(2-phenylethanoylamino)propyl]-1,2,3-triazol-1-yl]-4-oxidanyl-6-[(1~{R},2~{R})-1,2,3-tris(oxidanyl)propyl]oxane-2-carboxylic acid, (2~{R},4~{S},5~{R},6~{R})-5-acetamido-2-[4-[(2~{S})-3-[2-[(2~{S},3~{R},4~{R},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]ethylamino]-3-oxidanylidene-2-(2-phenylethanoylamino)propyl]-1,2,3-triazol-1-yl]-4-oxidanyl-6-[(1~{R},2~{R})-1,2,3-tris(oxidanyl)propyl]oxane-2-carboxylic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Heggelund, J.E, Mackenzie, A, Krengel, U. | | Deposit date: | 2016-09-29 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Towards new cholera prophylactics and treatment: Crystal structures of bacterial enterotoxins in complex with GM1 mimics.
Sci Rep, 7, 2017
|
|
4NDT
 
 | |
4NDV
 
 | |
4NDU
 
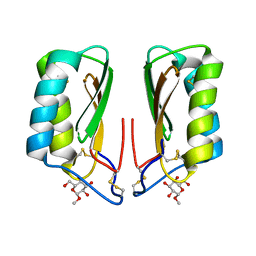 | |
4NDS
 
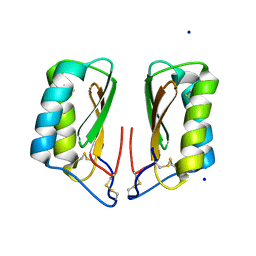 | |
1FNK
 
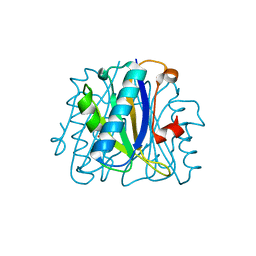 | | CRYSTAL STRUCTURE ANALYSIS OF CHORISMATE MUTASE MUTANT C88K/R90S | | Descriptor: | PROTEIN (CHORISMATE MUTASE) | | Authors: | Kast, P, Grisostomi, C, Chen, I.A, Li, S, Krengel, U, Xue, Y, Hilvert, D. | | Deposit date: | 2000-08-22 | | Release date: | 2000-10-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A strategically positioned cation is crucial for efficient catalysis by chorismate mutase.
J.Biol.Chem., 275, 2000
|
|
1FNJ
 
 | | CRYSTAL STRUCTURE ANALYSIS OF CHORISMATE MUTASE MUTANT C88S/R90K | | Descriptor: | PROTEIN (CHORISMATE MUTASE) | | Authors: | Kast, P, Grisostomi, C, Chen, I.A, Li, S, Krengel, U, Xue, Y, Hilvert, D. | | Deposit date: | 2000-08-22 | | Release date: | 2000-10-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A strategically positioned cation is crucial for efficient catalysis by chorismate mutase.
J.Biol.Chem., 275, 2000
|
|
6FFJ
 
 | |
1GR7
 
 | | Crystal structure of the double mutant Cys3Ser/Ser100Pro from Pseudomonas Aeruginosa at 1.8 A resolution | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Okvist, M, Bonander, N, Sandberg, A, Karlsson, B.G, Krengel, U, Xue, Y, Sjolin, L. | | Deposit date: | 2001-12-14 | | Release date: | 2002-05-16 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the double azurin mutant Cys3Ser/Ser100Pro from Pseudomonas aeruginosa at 1.8 A resolution: its folding-unfolding energy and unfolding kinetics.
Biochim.Biophys.Acta, 1596, 2002
|
|
1GZ9
 
 | | High-Resolution Crystal Structure of Erythrina cristagalli Lectin in Complex with 2'-alpha-L-Fucosyllactose | | Descriptor: | CALCIUM ION, ERYTHRINA CRISTA-GALLI LECTIN, MANGANESE (II) ION, ... | | Authors: | Svensson, C, Teneberg, S, Nilsson, C.L, Kjellberg, A, Schwarz, F.P, Sharon, N, Krengel, U. | | Deposit date: | 2002-05-17 | | Release date: | 2002-06-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-Resolution Crystal Structures of Erythrina Cristagalli Lectin in Complex with Lactose and 2'-Alpha-L-Fucosyllactose and Correlation with Thermodynamic Binding Data
J.Mol.Biol., 321, 2002
|
|
1GZC
 
 | |
5MPV
 
 | |
5MU9
 
 | | MOA-E-64 complex | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Agglutinin, ... | | Authors: | Cordara, G, Manna, D, Krengel, U. | | Deposit date: | 2017-01-12 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Family of Papain-Like Fungal Chimerolectins with Distinct Ca(2+)-Dependent Activation Mechanism.
Biochemistry, 56, 2017
|
|
5MUA
 
 | | PSL1a-E64 complex | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, ... | | Authors: | Cordara, G, Manna, D, Krengel, U. | | Deposit date: | 2017-01-12 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Family of Papain-Like Fungal Chimerolectins with Distinct Ca(2+)-Dependent Activation Mechanism.
Biochemistry, 56, 2017
|
|
