6GB9
 
 | |
5G2R
 
 | | Crystal structure of the Mo-insertase domain Cnx1E from Arabidopsis thaliana | | Descriptor: | GLYCEROL, MAGNESIUM ION, MOLYBDOPTERIN BIOSYNTHESIS PROTEIN CNX1, ... | | Authors: | Krausze, J, Saha, S, Probst, C, Kruse, T, Heinz, D.W, Mendel, R.R. | | Deposit date: | 2016-04-13 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Dimerization of the Plant Molybdenum Insertase Cnx1E is Required for Synthesis of the Molybdenum Cofactor.
Biochem.J., 474, 2017
|
|
5G2S
 
 | | Crystal structure of the Mo-insertase domain Cnx1E from Arabidopsis thaliana in complex with molybdate | | Descriptor: | GLYCEROL, MAGNESIUM ION, MOLYBDATE ION, ... | | Authors: | Krausze, J, Probst, C, Kruse, T, Heinz, D.W, Mendel, R.R. | | Deposit date: | 2016-04-13 | | Release date: | 2017-02-15 | | Method: | X-RAY DIFFRACTION (2.838 Å) | | Cite: | Dimerization of the Plant Molybdenum Insertase Cnx1E is Required for Synthesis of the Molybdenum Cofactor.
Biochem.J., 474, 2017
|
|
5OHX
 
 | | Structure of active cystathionine B-synthase from Apis mellifera | | Descriptor: | Cystathionine beta-synthase, PROTOPORPHYRIN IX CONTAINING FE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Gimenez-Mascarell, P, Majtan, T, Oyenarte, I, Ereno-Orbea, J, Majtan, J, Kraus, J.P, Klaudiny, J, Martinez-Cruz, L.A. | | Deposit date: | 2017-07-18 | | Release date: | 2018-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of cystathionine beta-synthase from honeybee Apis mellifera.
J. Struct. Biol., 202, 2018
|
|
1JBQ
 
 | | STRUCTURE OF HUMAN CYSTATHIONINE BETA-SYNTHASE: A UNIQUE PYRIDOXAL 5'-PHOSPHATE DEPENDENT HEMEPROTEIN | | Descriptor: | CYSTATHIONINE BETA-SYNTHASE, PROTOPORPHYRIN IX CONTAINING FE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Meier, M, Janosik, M, Kery, V, Kraus, J.P, Burkhard, P. | | Deposit date: | 2001-06-06 | | Release date: | 2001-08-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human cystathionine beta-synthase: a unique pyridoxal 5'-phosphate-dependent heme protein.
EMBO J., 20, 2001
|
|
4PCU
 
 | | Crystal structure of delta516-525 E201S human cystathionine beta-synthase with AdoMet | | Descriptor: | Cystathionine beta-synthase, PROTOPORPHYRIN IX CONTAINING FE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Ereno-Orbea, J, Majtan, T, Oyenarte, I, Kraus, J.P, Martinez-Cruz, L.A. | | Deposit date: | 2014-04-16 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.578 Å) | | Cite: | Structural insight into the molecular mechanism of allosteric activation of human cystathionine beta-synthase by S-adenosylmethionine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4L0D
 
 | | Crystal structure of delta516-525 human cystathionine beta-synthase containing C-terminal 6xHis-tag | | Descriptor: | Cystathionine beta-synthase, PROTOPORPHYRIN IX CONTAINING FE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ereno, J, Majtan, T, Oyenarte, I, Kraus, J.P, Martinez-Cruz, L.A. | | Deposit date: | 2013-05-31 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural basis of regulation and oligomerization of human cystathionine beta-synthase, the central enzyme of transsulfuration.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4L28
 
 | | Crystal structure of delta516-525 human cystathionine beta-synthase D444N mutant containing C-terminal 6xHis tag | | Descriptor: | Cystathionine beta-synthase, PROTOPORPHYRIN IX CONTAINING FE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ereno, J, Majtan, T, Oyenarte, I, Kraus, J.P, Martinez, L.A. | | Deposit date: | 2013-06-04 | | Release date: | 2013-09-18 | | Last modified: | 2013-10-16 | | Method: | X-RAY DIFFRACTION (2.626 Å) | | Cite: | Structural basis of regulation and oligomerization of human cystathionine beta-synthase, the central enzyme of transsulfuration.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4L27
 
 | | Crystal structure of delta1-39 and delta516-525 human cystathionine beta-synthase D444N mutant containing C-terminal 6xHis tag | | Descriptor: | Cystathionine beta-synthase, PROTOPORPHYRIN IX CONTAINING FE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ereno, J, Majtan, T, Oyenarte, I, Kraus, J.P, Martinez, L.A. | | Deposit date: | 2013-06-04 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.391 Å) | | Cite: | Structural basis of regulation and oligomerization of human cystathionine beta-synthase, the central enzyme of transsulfuration.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4L3V
 
 | | Crystal structure of delta516-525 human cystathionine beta-synthase | | Descriptor: | Cystathionine beta-synthase, PROTOPORPHYRIN IX CONTAINING FE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ereno, J, Majtan, T, Oyenarte, I, Kraus, J.P, Martinez, L.A. | | Deposit date: | 2013-06-07 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.628 Å) | | Cite: | Structural basis of regulation and oligomerization of human cystathionine beta-synthase, the central enzyme of transsulfuration.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3TIK
 
 | | Sterol 14-alpha demethylase (CYP51) from Trypanosoma brucei in complex with the tipifarnib derivative 6-((4-chlorophenyl)(methoxy)(1-methyl-1H-imidazol-5-yl)methyl)-4-(2,6-difluorophenyl)-1-methylquinolin-2(1H)-one | | Descriptor: | 6-[(R)-(4-chlorophenyl)(methoxy)(1-methyl-1H-imidazol-5-yl)methyl]-4-(2,6-difluorophenyl)-1-methylquinolin-2(1H)-one, PROTOPORPHYRIN IX CONTAINING FE, sterol 14-alpha demethylase (CYP51) | | Authors: | Hargrove, T.Y, Wawrzak, Z, Kraus, J.M, Gelb, M.H, Buckner, F.S, Waterman, M.R, Lepesheva, G.I. | | Deposit date: | 2011-08-20 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Pharmacological characterization, structural studies, and in vivo activities of anti-chagas disease lead compounds derived from tipifarnib.
Antimicrob.Agents Chemother., 56, 2012
|
|
8RZ6
 
 | | SeMet derivative structure of the condensation domain TomBC from the Tomaymycin non-ribosomal peptide synthetase | | Descriptor: | FORMIC ACID, GLYCEROL, POTASSIUM ION, ... | | Authors: | Karanth, M, Schmelz, S, Kirkpatrick, J, Krausze, J, Scrima, A, Carlomagno, T. | | Deposit date: | 2024-02-12 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The specificity of intermodular recognition in a prototypical nonribosomal peptide synthetase depends on an adaptor domain.
Sci Adv, 10, 2024
|
|
4UZC
 
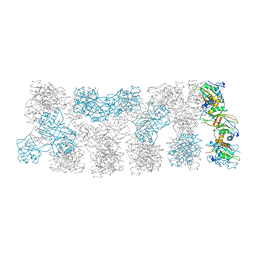 | |
4UZB
 
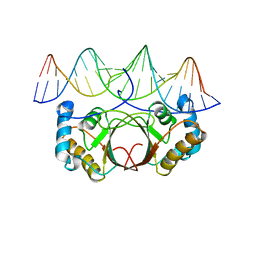 | | KSHV LANA (ORF73) C-terminal domain mutant bound to LBS1 DNA (R1039Q, R1040Q, K1055E, K1109A, D1110A, A1121E, K1138S, K1140D, K1141D) | | Descriptor: | LANA BINDING SITE 1 DNA, ORF 73 | | Authors: | Hellert, J, Krausze, J, Luhrs, T. | | Deposit date: | 2014-09-05 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.865 Å) | | Cite: | The 3D Structure of Kaposi Sarcoma Herpesvirus Lana C-Terminal Domain Bound to DNA.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8RH1
 
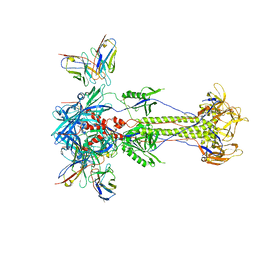 | | Trimeric HSV-2F gB ectodomain in postfusion conformation with three bound HDIT101 Fab molecules. | | Descriptor: | Envelope glycoprotein B, HDIT101 Fab heavy chain, HDIT101 Fab light chain | | Authors: | Kalbermatter, D, Seyfizadeh, N, Imhof, T, Ries, M, Mueller, C, Jenner, L, Blumenschein, E, Yendrzheyevskiy, A, Moog, K, Eckert, D, Engel, R, Diebolder, P, Chami, M, Krauss, J, Schaller, T, Arndt, M. | | Deposit date: | 2023-12-14 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Development of a highly effective combination monoclonal antibody therapy against Herpes simplex virus.
J.Biomed.Sci., 31, 2024
|
|
8RH2
 
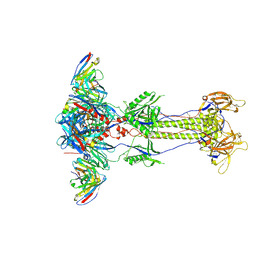 | | Trimeric HSV-2G gB ectodomain in postfusion conformation with three bound HDIT102 Fab molecules. | | Descriptor: | Envelope glycoprotein B, HDIT102 Fab heavy chain, HDIT102 Fab light chain | | Authors: | Kalbermatter, D, Seyfizadeh, N, Imhof, T, Ries, M, Mueller, C, Jenner, L, Blumenschein, E, Yendrzheyevskiy, A, Moog, K, Eckert, D, Engel, R, Diebolder, P, Chami, M, Krauss, J, Schaller, T, Arndt, M. | | Deposit date: | 2023-12-14 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Development of a highly effective combination monoclonal antibody therapy against Herpes simplex virus.
J.Biomed.Sci., 31, 2024
|
|
8RGZ
 
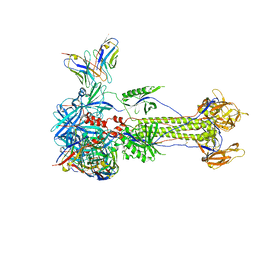 | | Trimeric HSV-1F gB ectodomain in postfusion conformation with three bound HDIT101 Fab molecules. | | Descriptor: | Envelope glycoprotein B, HDIT101 Fab heavy chain, HDIT101 Fab light chain | | Authors: | Kalbermatter, D, Seyfizadeh, N, Imhof, T, Ries, M, Mueller, C, Jenner, L, Blumenschein, E, Yendrzheyevskiy, A, Moog, K, Eckert, D, Engel, R, Diebolder, P, Chami, M, Krauss, J, Schaller, T, Arndt, M. | | Deposit date: | 2023-12-14 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Development of a highly effective combination monoclonal antibody therapy against Herpes simplex virus.
J.Biomed.Sci., 31, 2024
|
|
8RH0
 
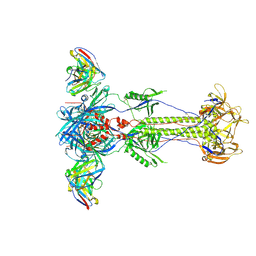 | | Trimeric HSV-1F gB ectodomain in postfusion conformation with three bound HDIT102 Fab molecules. | | Descriptor: | Envelope glycoprotein B, HDIT102 Fab heavy chain | | Authors: | Kalbermatter, D, Seyfizadeh, N, Imhof, T, Ries, M, Mueller, C, Jenner, L, Blumenschein, E, Yendrzheyevskiy, A, Moog, K, Eckert, D, Engel, R, Diebolder, P, Chami, M, Krauss, J, Schaller, T, Arndt, M. | | Deposit date: | 2023-12-14 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Development of a highly effective combination monoclonal antibody therapy against Herpes simplex virus.
J.Biomed.Sci., 31, 2024
|
|
8QNF
 
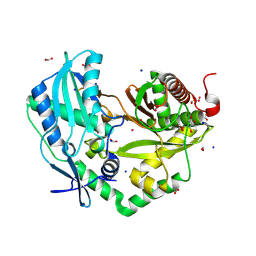 | | Crystal structure of the Condensation domain TomBC from the Tomaymycin non-ribosomal peptide synthetase | | Descriptor: | Condensation domain TomBC from the Tomaymycin non-ribosomal peptide synthetase, FORMIC ACID, GLYCEROL, ... | | Authors: | Karanth, M, Schmelz, S, Kirkpatrick, J, Krausze, J, Scrima, A, Carlomagno, T. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The specificity of intermodular recognition in a prototypical nonribosomal peptide synthetase depends on an adaptor domain.
Sci Adv, 10, 2024
|
|
4A4V
 
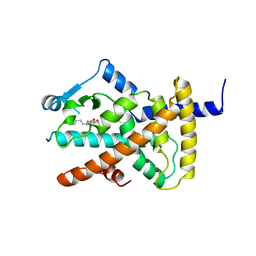 | | Ligand binding domain of human PPAR gamma in complex with amorfrutin 2 | | Descriptor: | AMORFRUTIN 2, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | de Groot, J.C, Weidner, C, Krausze, J, Kawamoto, K, Schroeder, F.C, Sauer, S, Buessow, K. | | Deposit date: | 2011-10-20 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of Amorfrutins Bound to the Peroxisome Proliferator-Activated Receptor Gamma.
J.Med.Chem., 56, 2013
|
|
4A4W
 
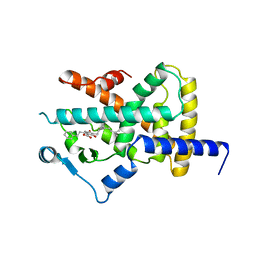 | | Ligand binding domain of human PPAR gamma in complex with amorfrutin B | | Descriptor: | AMORFRUTIN B, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | de Groot, J.C, Weidner, C, Krausze, J, Kawamoto, K, Schroeder, F.C, Sauer, S, Buessow, K. | | Deposit date: | 2011-10-20 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of Amorfrutins Bound to the Peroxisome Proliferator-Activated Receptor Gamma.
J.Med.Chem., 56, 2013
|
|
2YPZ
 
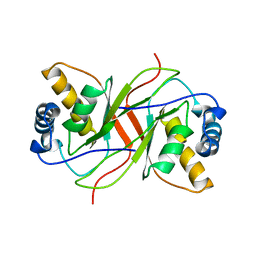 | |
2YBO
 
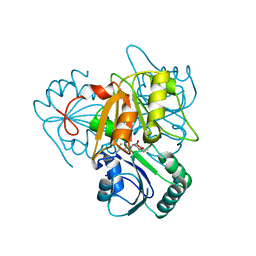 | | The x-ray structure of the SAM-dependent uroporphyrinogen III methyltransferase NirE from Pseudomonas aeruginosa in complex with SAH | | Descriptor: | METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Storbeck, S, Saha, S, Krausze, J, Klink, B.U, Heinz, D.W, Layer, G. | | Deposit date: | 2011-03-08 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Heme D1 Biosynthesis Enzyme Nire in Complex with its Substrate Reveals New Insights Into the Catalytic Mechanism of S-Adenosyl-L-Methionine-Dependent Uroporphyrinogen III Methyltransferases.
J.Biol.Chem., 286, 2011
|
|
2YBQ
 
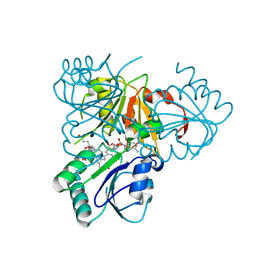 | | The x-ray structure of the SAM-dependent uroporphyrinogen III methyltransferase NirE from Pseudomonas aeruginosa in complex with SAH and uroporphyrinogen III | | Descriptor: | METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE, UROPORPHYRINOGEN III | | Authors: | Storbeck, S, Saha, S, Krausze, J, Klink, B.U, Heinz, D.W, Layer, G. | | Deposit date: | 2011-03-09 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Heme D1 Biosynthesis Enzyme Nire in Complex with its Substrate Reveals New Insights Into the Catalytic Mechanism of S-Adenosyl-L-Methionine-Dependent Uroporphyrinogen III Methyltransferases.
J.Biol.Chem., 286, 2011
|
|
2YQ0
 
 | |
