3B7V
 
 | | HIV-1 protease complexed with gem-diol-amine tetrahedral intermediate NLLTQI | | Descriptor: | CHLORIDE ION, GLYCEROL, Protease, ... | | Authors: | Kovalevsky, A.Y, Chumanevich, A.A, Weber, I.T. | | Deposit date: | 2007-10-31 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Caught in the Act: The 1.5 A Resolution Crystal Structures of the HIV-1 Protease and the I54V Mutant Reveal a Tetrahedral Reaction Intermediate.
Biochemistry, 46, 2007
|
|
4DVO
 
 | | Room-temperature joint X-ray/neutron structure of D-xylose isomerase in complex with 2Ni2+ and per-deuterated D-sorbitol at pH 5.9 | | Descriptor: | NICKEL (II) ION, Xylose isomerase, sorbitol | | Authors: | Kovalevsky, A.Y, Hanson, L, Langan, P. | | Deposit date: | 2012-02-23 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Inhibition of D-xylose isomerase by polyols: atomic details by joint X-ray/neutron crystallography.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4S2D
 
 | | Joint X-ray/neutron structure of Trichoderma reesei xylanase II in complex with MES at pH 5.7 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Kovalevsky, A.Y, Wan, Q, Langan, P. | | Deposit date: | 2015-01-20 | | Release date: | 2015-09-23 | | Last modified: | 2024-11-27 | | Method: | NEUTRON DIFFRACTION (1.6 Å), X-RAY DIFFRACTION | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7LB7
 
 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (3CL Mpro) in complex with Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A.Y, Kneller, D.W, Coates, L. | | Deposit date: | 2021-01-07 | | Release date: | 2021-01-20 | | Last modified: | 2024-11-13 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Direct Observation of Protonation State Modulation in SARS-CoV-2 Main Protease upon Inhibitor Binding with Neutron Crystallography.
J.Med.Chem., 64, 2021
|
|
4DH8
 
 | | Room temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, AMP-PNP and IP20 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4JEC
 
 | | Joint neutron and X-ray structure of per-deuterated HIV-1 protease in complex with clinical inhibitor amprenavir | | Descriptor: | CHLORIDE ION, HIV-1 protease, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | Authors: | Kovalevsky, A.Y, Weber, I.T, Langan, P. | | Deposit date: | 2013-02-26 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (2.01 Å), X-RAY DIFFRACTION | | Cite: | Joint X-ray/Neutron Crystallographic Study of HIV-1 Protease with Clinical Inhibitor Amprenavir: Insights for Drug Design.
J.Med.Chem., 56, 2013
|
|
3OXC
 
 | | Wild Type HIV-1 Protease with Antiviral Drug Saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, FORMIC ACID, Protease, ... | | Authors: | Kovalevsky, A.Y, Wang, Y.-F, Tie, Y, Weber, I.T. | | Deposit date: | 2010-09-21 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir
Proteins, 67, 2007
|
|
4DUO
 
 | | Room-temperature X-ray structure of D-Xylose Isomerase in complex with 2Mg2+ ions and xylitol at pH 7.7 | | Descriptor: | MAGNESIUM ION, Xylitol, Xylose isomerase | | Authors: | Kovalevsky, A.Y, Hanson, L, Langan, P. | | Deposit date: | 2012-02-22 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of D-xylose isomerase by polyols: atomic details by joint X-ray/neutron crystallography.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5EBJ
 
 | | Joint X-ray/neutron structure of reversibly photoswitching chromogenic protein, Dathail | | Descriptor: | photoswitching chromogenic protein | | Authors: | Kovalevsky, A.Y, Langan, P.S, Bradbury, A.R.M. | | Deposit date: | 2015-10-19 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-23 | | Method: | NEUTRON DIFFRACTION (2.5 Å), X-RAY DIFFRACTION | | Cite: | Evolution and characterization of a new reversibly photoswitching chromogenic protein, Dathail.
J.Mol.Biol., 428, 2016
|
|
5E5K
 
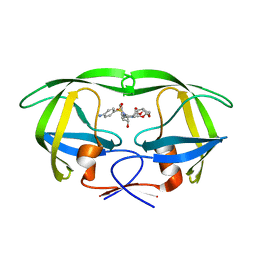 | | Joint X-ray/neutron structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with darunavir at pH 4.3 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, HIV-1 protease | | Authors: | Kovalevsky, A.Y, Das, A. | | Deposit date: | 2015-10-08 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.75 Å), X-RAY DIFFRACTION | | Cite: | Long-Range Electrostatics-Induced Two-Proton Transfer Captured by Neutron Crystallography in an Enzyme Catalytic Site.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5E5J
 
 | | Joint X-ray/neutron structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with darunavir at pH 6.0 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Kovalevsky, A.Y, Gerlits, O.O. | | Deposit date: | 2015-10-08 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | Cite: | Long-Range Electrostatics-Induced Two-Proton Transfer Captured by Neutron Crystallography in an Enzyme Catalytic Site.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
3QYS
 
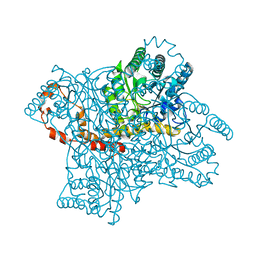 | |
3QZA
 
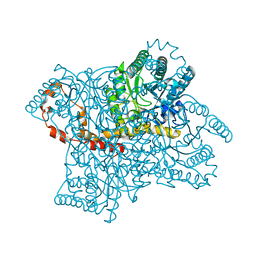 | |
4DH5
 
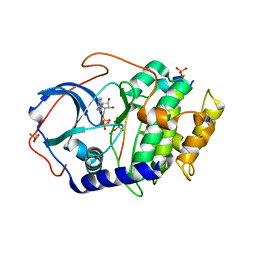 | | Room temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, ADP, Phosphate, and IP20 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DH3
 
 | | Low temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, ATP and IP20 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DH7
 
 | | Low temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with high Mg2+, AMP-PNP and IP20' | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DH1
 
 | | Low temperature X-ray structure of cAMP dependent Protein Kinase A catalytic subunit with low Mg2+, ATP and IP20 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2012-01-27 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Low- and room-temperature X-ray structures of protein kinase A ternary complexes shed new light on its activity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
8DT4
 
 | | X-ray structure of human acetylcholinesterase ternary complex with paraoxon and oxime MMB4 (POX-hAChE-MMB4) | | Descriptor: | 1,1'-methylenebis{4-[(E)-(hydroxyimino)methyl]pyridin-1-ium}, Acetylcholinesterase, DIETHYL PHOSPHONATE, ... | | Authors: | Kovalevsky, A.Y, Gerlits, O, Radic, Z. | | Deposit date: | 2022-07-25 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and dynamic effects of paraoxon binding to human acetylcholinesterase by X-ray crystallography and inelastic neutron scattering.
Structure, 30, 2022
|
|
8DT2
 
 | | X-ray structure of human acetylcholinesterase inhibited by paraoxon (POX-hAChE) | | Descriptor: | Acetylcholinesterase, DIETHYL PHOSPHONATE, DIMETHYL SULFOXIDE, ... | | Authors: | Kovalevsky, A.Y, Gerlits, O, Radic, Z. | | Deposit date: | 2022-07-25 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Structural and dynamic effects of paraoxon binding to human acetylcholinesterase by X-ray crystallography and inelastic neutron scattering.
Structure, 30, 2022
|
|
8DT7
 
 | | X-ray structure of human acetylcholinesterase in complex with oxime MMB4 (hAChE-MMB4) | | Descriptor: | 1,1'-methylenebis{4-[(E)-(hydroxyimino)methyl]pyridin-1-ium}, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Kovalevsky, A.Y, Gerlits, O, Radic, Z. | | Deposit date: | 2022-07-25 | | Release date: | 2022-11-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.207 Å) | | Cite: | Structural and dynamic effects of paraoxon binding to human acetylcholinesterase by X-ray crystallography and inelastic neutron scattering.
Structure, 30, 2022
|
|
8DT5
 
 | | X-ray structure of human acetylcholinesterase ternary complex with paraoxon and oxime RS170B (POX-hAChE-RS170B) | | Descriptor: | 4-carbamoyl-1-(3-{2-[(E)-(hydroxyimino)methyl]-1H-imidazol-1-yl}propyl)pyridin-1-ium, Acetylcholinesterase, DIETHYL PHOSPHONATE, ... | | Authors: | Kovalevsky, A.Y, Gerlits, O, Radic, Z. | | Deposit date: | 2022-07-25 | | Release date: | 2022-11-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and dynamic effects of paraoxon binding to human acetylcholinesterase by X-ray crystallography and inelastic neutron scattering.
Structure, 30, 2022
|
|
8DL9
 
 | |
8DLB
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease in complex with compound Z2799209083 | | Descriptor: | 1-[(5S)-5-(3,4-dimethoxyphenyl)-3-phenyl-4,5-dihydro-1H-pyrazol-1-yl]ethan-1-one, 3C-like proteinase | | Authors: | Kovalevsky, A.Y, Coates, L, Kneller, D.W. | | Deposit date: | 2022-07-07 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | AI-Accelerated Design of Targeted Covalent Inhibitors for SARS-CoV-2.
J.Chem.Inf.Model., 63, 2023
|
|
3KBS
 
 | |
3KBV
 
 | |
