6D54
 
 | |
6E21
 
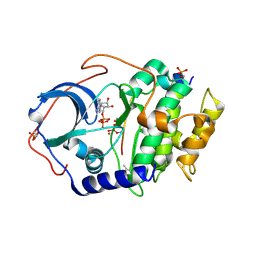 | | Joint X-ray/neutron structure of PKAc with products Sr2-ADP and phosphorylated peptide SP20 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, STRONTIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Kovalevsky, A, Gerlits, O.O, Taylor, S. | | Deposit date: | 2018-07-10 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Zooming in on protons: Neutron structure of protein kinase A trapped in a product complex.
Sci Adv, 5, 2019
|
|
3OXC
 
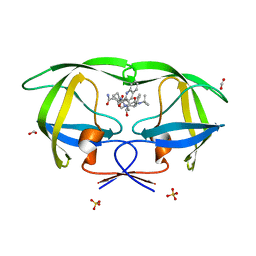 | | Wild Type HIV-1 Protease with Antiviral Drug Saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, FORMIC ACID, Protease, ... | | Authors: | Kovalevsky, A.Y, Wang, Y.-F, Tie, Y, Weber, I.T. | | Deposit date: | 2010-09-21 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir
Proteins, 67, 2007
|
|
2G69
 
 | |
5E5K
 
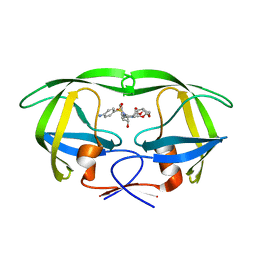 | | Joint X-ray/neutron structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with darunavir at pH 4.3 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, HIV-1 protease | | Authors: | Kovalevsky, A.Y, Das, A. | | Deposit date: | 2015-10-08 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.75 Å), X-RAY DIFFRACTION | | Cite: | Long-Range Electrostatics-Induced Two-Proton Transfer Captured by Neutron Crystallography in an Enzyme Catalytic Site.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5E5J
 
 | | Joint X-ray/neutron structure of HIV-1 protease triple mutant (V32I,I47V,V82I) with darunavir at pH 6.0 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Kovalevsky, A.Y, Gerlits, O.O. | | Deposit date: | 2015-10-08 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | Cite: | Long-Range Electrostatics-Induced Two-Proton Transfer Captured by Neutron Crystallography in an Enzyme Catalytic Site.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5EBJ
 
 | | Joint X-ray/neutron structure of reversibly photoswitching chromogenic protein, Dathail | | Descriptor: | photoswitching chromogenic protein | | Authors: | Kovalevsky, A.Y, Langan, P.S, Bradbury, A.R.M. | | Deposit date: | 2015-10-19 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-23 | | Method: | NEUTRON DIFFRACTION (2.5 Å), X-RAY DIFFRACTION | | Cite: | Evolution and characterization of a new reversibly photoswitching chromogenic protein, Dathail.
J.Mol.Biol., 428, 2016
|
|
2F81
 
 | | HIV-1 Protease mutant L90M complexed with inhibitor TMC114 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2005-12-01 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Effectiveness of Nonpeptide Clinical Inhibitor TMC-114 on HIV-1 Protease with Highly Drug Resistant Mutations D30N, I50V, and L90M.
J.Med.Chem., 49, 2006
|
|
7SI9
 
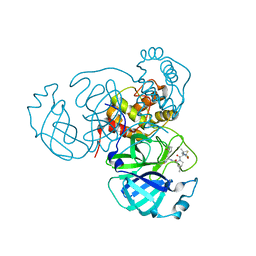 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with PF-07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-10-12 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
2HB3
 
 | | Wild-type HIV-1 Protease in complex with potent inhibitor GRL06579 | | Descriptor: | (3AS,5R,6AR)-HEXAHYDRO-2H-CYCLOPENTA[B]FURAN-5-YL (2S,3S)-3-HYDROXY-4-(4-(HYDROXYMETHYL)-N-ISOBUTYLPHENYLSULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2006-06-13 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-Based Design of Novel HIV-1 Protease Inhibitors To Combat Drug Resistance.
J.Med.Chem., 49, 2006
|
|
7TEH
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-2 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-05 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7TDU
 
 | | Joint X-ray/neutron structure of SARS-CoV-2 main protease (3CL Mpro) in complex with BBH-1 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxo(1-~2~H)pyrrolidin-3-yl]propan-2-yl}-3-{N-[tert-butyl(~2~H)carbamoyl]-3-methyl-L-(N-~2~H)valyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-(~2~H)carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-03 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | NEUTRON DIFFRACTION (1.85 Å), X-RAY DIFFRACTION | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
7TFR
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with NBH-2 | | Descriptor: | (1R,2S,5S)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-({1-[(2-methylpropane-2-sulfonyl)methyl]cyclohexyl}carbamoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-07 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
3QYS
 
 | |
3QZA
 
 | |
3CWH
 
 | | D-xylose Isomerase in complex with linear product, per-deuterated xylulose | | Descriptor: | D-XYLULOSE, HYDROXIDE ION, MAGNESIUM ION, ... | | Authors: | Kovalevsky, A.Y, Langan, P, Glusker, J.P. | | Deposit date: | 2008-04-21 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | Hydrogen location in stages of an enzyme-catalyzed reaction: time-of-flight neutron structure of D-xylose isomerase with bound D-xylulose
Biochemistry, 47, 2008
|
|
4QEE
 
 | |
4QDW
 
 | | Joint X-ray and neutron structure of Streptomyces rubiginosus D-xylose isomerase in complex with two Ni2+ ions and linear L-arabinose | | Descriptor: | L-arabinose, NICKEL (II) ION, Xylose isomerase | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2014-05-14 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | Cite: | L-Arabinose Binding, Isomerization, and Epimerization by D-Xylose Isomerase: X-Ray/Neutron Crystallographic and Molecular Simulation Study.
Structure, 22, 2014
|
|
4QE5
 
 | |
4QE4
 
 | |
4QDP
 
 | | Joint X-ray and neutron structure of Streptomyces rubiginosus D-xylose isomerase in complex with two Cd2+ ions and cyclic beta-L-arabinose | | Descriptor: | CADMIUM ION, Xylose isomerase, beta-L-arabinopyranose | | Authors: | Kovalevsky, A.Y, Langan, P. | | Deposit date: | 2014-05-14 | | Release date: | 2014-09-03 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | L-Arabinose Binding, Isomerization, and Epimerization by D-Xylose Isomerase: X-Ray/Neutron Crystallographic and Molecular Simulation Study.
Structure, 22, 2014
|
|
4QEH
 
 | |
4QE1
 
 | |
3EBZ
 
 | | High Resolution HIV-2 Protease Structure in Complex with Clinical Drug Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, IMIDAZOLE, ... | | Authors: | Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2008-08-28 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural evidence for effectiveness of darunavir and two related antiviral inhibitors against HIV-2 protease
J.Mol.Biol., 384, 2008
|
|
3ECG
 
 | | High Resolution HIV-2 Protease Structure in Complex with Antiviral Inhibitor GRL-98065 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(2S,3R)-3-HYDROXY-4-(N-ISOBUTYLBENZO[D][1,3]DIOXOLE-5-SULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, CHLORIDE ION, IMIDAZOLE, ... | | Authors: | Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2008-08-29 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structural evidence for effectiveness of darunavir and two related antiviral inhibitors against HIV-2 protease
J.Mol.Biol., 384, 2008
|
|
