3IVK
 
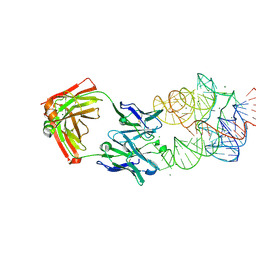 | | Crystal Structure of the Catalytic Core of an RNA Polymerase Ribozyme Complexed with an Antigen Binding Antibody Fragment | | Descriptor: | CADMIUM ION, CHLORIDE ION, Fab heavy chain, ... | | Authors: | Koldobskaya, Y, Duguid, E.M, Shechner, D.M, Koide, S, Kossiakoff, A.A, Bartel, D.P, Piccirilli, J.A. | | Deposit date: | 2009-09-01 | | Release date: | 2010-03-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the catalytic core of an RNA-polymerase ribozyme.
Science, 326, 2009
|
|
3E8Y
 
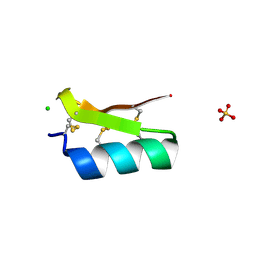 | | Xray structure of scorpion toxin BmBKTx1 | | Descriptor: | CHLORIDE ION, Potassium channel toxin alpha-KTx 19.1, SULFATE ION | | Authors: | Mandal, K, Pentelute, B.L, Tereshko, V, Kossiakoff, A.A, Kent, S.B.H. | | Deposit date: | 2008-08-20 | | Release date: | 2009-02-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-ray structure of native scorpion toxin BmBKTx1 by racemic protein crystallography using direct methods.
J.Am.Chem.Soc., 131, 2009
|
|
2FXU
 
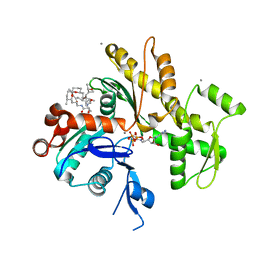 | | X-ray Structure of Bistramide A- Actin Complex at 1.35 A resolution. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Rizvi, S.A, Tereshko, V, Kossiakoff, A.A, Kozmin, S.A. | | Deposit date: | 2006-02-06 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of bistramide a-actin complex at a 1.35 A resolution
J.Am.Chem.Soc., 128, 2006
|
|
3EFD
 
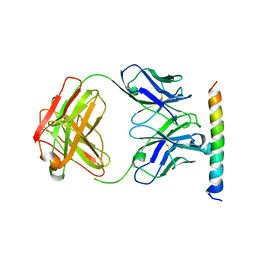 | | The crystal structure of the cytoplasmic domain of KcsA | | Descriptor: | FabH, FabL, KcsA | | Authors: | Uysal, S, Vasquez, V, Tereshko, V, Esaki, K, Fellouse, F.A, Sidhu, S.S, Koide, S, Perozo, E, Kossiakoff, A. | | Deposit date: | 2008-09-08 | | Release date: | 2009-04-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of full-length KcsA in its closed conformation.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3EFF
 
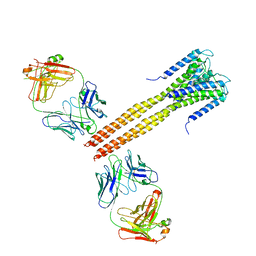 | | The Crystal Structure of Full-Length KcsA in its Closed Conformation | | Descriptor: | FAB, Voltage-gated potassium channel | | Authors: | Uysal, S, Vasquez, V, Tereshko, T, Esaki, K, Fellouse, F.A, Sidhu, S.S, Koide, S, Perozo, E, Kossiakoff, A. | | Deposit date: | 2008-09-08 | | Release date: | 2009-04-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal structure of full-length KcsA in its closed conformation.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6CBV
 
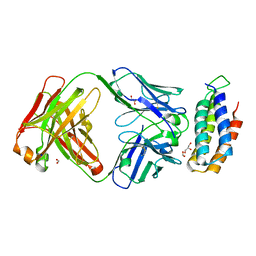 | | Crystal structure of BRIL bound to an affinity matured synthetic antibody. | | Descriptor: | BRIL, FORMIC ACID, GLYCEROL, ... | | Authors: | Mukherjee, S, Skrobek, B, Kossiakoff, A.A. | | Deposit date: | 2018-02-05 | | Release date: | 2019-02-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.872 Å) | | Cite: | Synthetic antibodies against BRIL as universal fiducial marks for single-particle cryoEM structure determination of membrane proteins.
Nat Commun, 11, 2020
|
|
1Z7C
 
 | |
3BOI
 
 | | Snow Flea Antifreeze Protein Racemate | | Descriptor: | 6.5 kDa glycine-rich antifreeze protein | | Authors: | Pentelute, B.L, Kent, S.B.H, Gates, Z.P, Tereshko, V, Kossiakoff, A.A, Kurutz, J, Dashnau, J, Vaderkooi, J.M. | | Deposit date: | 2007-12-17 | | Release date: | 2008-09-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | X-ray structure of snow flea antifreeze protein determined by racemic crystallization of synthetic protein enantiomers
J.Am.Chem.Soc., 130, 2008
|
|
6CMO
 
 | | Rhodopsin-Gi complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab Heavy chain, Fab light chain, ... | | Authors: | Kang, Y, Kuybeda, O, de Waal, P.W, Mukherjee, S, Van Eps, N, Dutka, P, Zhou, X.E, Bartesaghi, A, Erramilli, S, Morizumi, T, Gu, X, Yin, Y, Liu, P, Jiang, Y, Meng, X, Zhao, G, Melcher, K, Earnst, O.P, Kossiakoff, A.A, Subramaniam, S, Xu, H.E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of human rhodopsin bound to an inhibitory G protein.
Nature, 558, 2018
|
|
5W2B
 
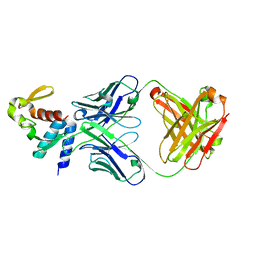 | | Crystal structure of C-terminal domain of Ebola (Reston) nucleoprotein in complex with Fab fragment | | Descriptor: | Fab Heavy Chain, Fab Light Chain, Nucleoprotein | | Authors: | Radwanska, M.J, Derewenda, U, Kossiakoff, A.A, Derewenda, Z.S. | | Deposit date: | 2017-06-06 | | Release date: | 2018-06-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure of the C-terminal domain of the nucleoprotein from the Bundibugyo strain of the Ebola virus in complex with a pan-specific synthetic Fab.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
1Y3T
 
 | | Crystal structure of YxaG, a dioxygenase from Bacillus subtilis | | Descriptor: | FE (III) ION, Hypothetical protein yxaG | | Authors: | Gopal, B, Madan, L.L, Betz, S.F, Kossiakoff, A.A. | | Deposit date: | 2004-11-26 | | Release date: | 2005-01-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of a Quercetin 2,3-Dioxygenase from Bacillus subtilis Suggests Modulation of Enzyme Activity by a Change in the Metal Ion at the Active Site(s)
Biochemistry, 44, 2005
|
|
1YJ1
 
 | | X-ray Crystal Structure of a Chemically Synthesized [D-Gln35]Ubiquitin | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ubiquitin | | Authors: | Bang, D, Makhatadze, G.I, Tereshko, V, Kossiakoff, A.A, Kent, S.B. | | Deposit date: | 2005-01-13 | | Release date: | 2005-06-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | X-ray Crystal Structure of a Chemically Synthesized [D-Gln35]Ubiquitin
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
1YIW
 
 | | X-ray Crystal Structure of a Chemically Synthesized Ubiquitin | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ubiquitin | | Authors: | Bang, D, Makhatadze, G.I, Tereshko, V, Kossiakoff, A.A, Kent, S.B. | | Deposit date: | 2005-01-13 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | X-ray Crystal Structure of a Chemically Synthesized [D-Gln35]Ubiquitin
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
6B6Z
 
 | | Crystal structure of the Apo Antibody fragment (Fab) raised against C-terminal domain of Ebola nucleoprotein (EBOV, TAFV, BDBV strains) | | Descriptor: | Apo Fab Heavy Chain, Apo Fab Light Chain, ZINC ION | | Authors: | Radwanska, M.J, Derewenda, U, Kossiakoff, A.A, Derewenda, Z.S. | | Deposit date: | 2017-10-03 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | Crystal structure of the Apo Antibody fragment (Fab) raised against C-terminal domain of Ebola nucleoprotein (EBOV, TAFV, BDBV strains)
To Be Published
|
|
1AAL
 
 | |
5CWS
 
 | | Crystal structure of the intact Chaetomium thermophilum Nsp1-Nup49-Nup57 channel nucleoporin heterotrimer bound to its Nic96 nuclear pore complex attachment site | | Descriptor: | Nucleoporin NIC96, Nucleoporin NSP1, Nucleoporin NUP49, ... | | Authors: | Bley, C.J, Petrovic, S, Paduch, M, Lu, V, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
6AZ2
 
 | | Crystal structure of Asf1-Fab 12E complex | | Descriptor: | Fab Heavy Chain, Fab Light Chain, Histone chaperone ASF1 | | Authors: | Bailey, L.J, Kossiakoff, A.A. | | Deposit date: | 2017-09-09 | | Release date: | 2018-01-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.477 Å) | | Cite: | Locking the Elbow: Improved Antibody Fab Fragments as Chaperones for Structure Determination.
J. Mol. Biol., 430, 2018
|
|
6AYZ
 
 | | Crystal structure of Asf1-Fab 12E complex | | Descriptor: | Fab Heavy Chain, Fab Light Chain, Histone chaperone ASF1 | | Authors: | Bailey, L.J, Kossiakoff, A.A. | | Deposit date: | 2017-09-08 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Locking the Elbow: Improved Antibody Fab Fragments as Chaperones for Structure Determination.
J. Mol. Biol., 430, 2018
|
|
5CWV
 
 | | Crystal structure of Chaetomium thermophilum Nup192 TAIL domain | | Descriptor: | Nucleoporin NUP192 | | Authors: | Stuwe, T, Bley, C.J, Thierbach, K, Petrovic, S, Schilbach, S, Mayo, D.J, Perriches, T, Rundlet, E.J, Jeon, Y.E, Collins, L.N, Lin, D.H, Paduch, M, Koide, A, Lu, V, Fischer, J, Hurt, E, Koide, S, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.155 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
5CWW
 
 | | Crystal structure of the Chaetomium thermophilum heterotrimeric Nup82 NTD-Nup159 TAIL-Nup145N APD complex | | Descriptor: | Nucleoporin NUP145N, Nucleoporin NUP159, Nucleoporin NUP82 | | Authors: | Stuwe, T, Bley, C.J, Thierbach, K, Petrovic, S, Schilbach, S, Mayo, D.J, Perriches, T, Rundlet, E.J, Jeon, Y.E, Collins, L.N, Lin, D.H, Paduch, M, Koide, A, Lu, V, Fischer, J, Hurt, E, Koide, S, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
1CA0
 
 | | BOVINE CHYMOTRYPSIN COMPLEXED TO APPI | | Descriptor: | BOVINE CHYMOTRYPSIN, PROTEASE INHIBITOR DOMAIN OF ALZHEIMER'S AMYLOID BETA-PROTEIN PRECURSOR | | Authors: | Scheidig, A.J, Kossiakoff, A.A. | | Deposit date: | 1997-01-23 | | Release date: | 1997-07-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of bovine chymotrypsin and trypsin complexed to the inhibitor domain of Alzheimer's amyloid beta-protein precursor (APPI) and basic pancreatic trypsin inhibitor (BPTI): engineering of inhibitors with altered specificities.
Protein Sci., 6, 1997
|
|
1CBW
 
 | | BOVINE CHYMOTRYPSIN COMPLEXED TO BPTI | | Descriptor: | BOVINE CHYMOTRYPSIN, BPTI, SULFATE ION | | Authors: | Hynes, T.R, Scheidig, A.J, Kossiakoff, A.A. | | Deposit date: | 1996-12-22 | | Release date: | 1997-07-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of bovine chymotrypsin and trypsin complexed to the inhibitor domain of Alzheimer's amyloid beta-protein precursor (APPI) and basic pancreatic trypsin inhibitor (BPTI): engineering of inhibitors with altered specificities.
Protein Sci., 6, 1997
|
|
6BF9
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain | | Descriptor: | Fab H11-E heavy chain, Fab H11-E light chain, Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-26 | | Release date: | 2018-02-07 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
6BF7
 
 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain | | Descriptor: | Fab H11-E heavy chain, Fab H11-E light chain, Insulin-degrading enzyme | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-26 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
1S6Y
 
 | | 2.3A crystal structure of phospho-beta-glucosidase | | Descriptor: | 6-phospho-beta-glucosidase | | Authors: | Tereshko, V, Dementieva, I, Kim, Y, Collat, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-01-28 | | Release date: | 2004-05-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | 2.3A CRYSTAL STRUCTURE OF PHOSPHO-BETA-GLUCOSIDASE, licH Gene Product from BACILLUS STEAROTHERMOPHILUS
To be Published
|
|
