2DZ7
 
 | | DNA Octaplex Formation with an I-Motif of A-Quartets: The Revised Crystal Structure of d(GCGAAAGC) | | Descriptor: | DNA (5'-D(*DGP*DCP*DGP*DAP*DAP*DAP*DGP*DC)-3'), MAGNESIUM ION | | Authors: | Sato, Y, Mitomi, K, Sumani, T, Kondo, J, Takenaka, A. | | Deposit date: | 2006-09-21 | | Release date: | 2007-08-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DNA octaplex formation with an I-motif of water-mediated A-quartets: reinterpretation of the crystal structure of d(GCGAAAGC)
J.Biochem.(Tokyo), 140, 2006
|
|
4K32
 
 | | Crystal structure of geneticin bound to the leishmanial rRNA A-site | | Descriptor: | GENETICIN, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*GP*UP*UP*CP*CP*GP*GP*AP*AP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Shalev, M, Kondo, J, Adir, N, Baasov, T. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of the molecular attributes required for aminoglycoside activity against Leishmania.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4K31
 
 | | Crystal structure of apramycin bound to the leishmanial rRNA A-site | | Descriptor: | APRAMYCIN, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*GP*UP*UP*CP*CP*GP*GP*AP*AP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Shalev, M, Kondo, J, Adir, N, Baasov, T. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.415 Å) | | Cite: | Identification of the molecular attributes required for aminoglycoside activity against Leishmania.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1UE2
 
 | | Crystal structure of d(GC38GAAAGCT) | | Descriptor: | 5'-D(*GP*(C38)P*GP*AP*AP*AP*GP*CP*T)-3', CHLORIDE ION, COBALT HEXAMMINE(III), ... | | Authors: | Sunami, T, Kondo, J, Hirao, I, Watanaba, K, Miura, K, Takenaka, A. | | Deposit date: | 2003-05-08 | | Release date: | 2004-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of d(GCGAAAGC) (hexagonal form): a base-intercalated duplex as a stable structure.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1UB8
 
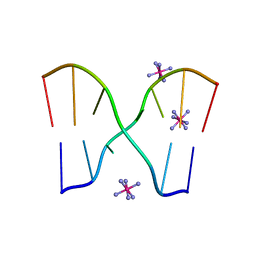 | | Crystal structure of d(GCGAAGC), bending duplex with a bulge-in residue | | Descriptor: | 5'-D(*GP*CP*GP*AP*AP*GP*C)-3', COBALT HEXAMMINE(III) | | Authors: | Sunami, T, Kondo, J, Hirao, I, Watanabe, K, Miura, K, Takenaka, A. | | Deposit date: | 2003-03-31 | | Release date: | 2004-03-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of d(GCGAAGC) and d(GCGAAAGC) (tetragonal form): a switching of partners of the sheared G.A pairs to form a functional G.AxA.G crossing.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5XWG
 
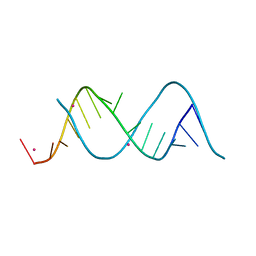 | |
3S4P
 
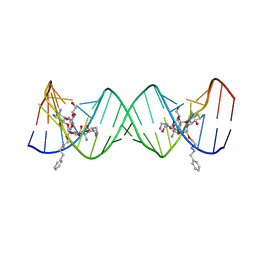 | | Crystal structure of the bacterial ribosomal decoding site complexed with an amphiphilic paromomycin O2''-ether analogue | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-2-O-{2-[(2-phenylethyl)amino]ethyl}-beta-D-ribofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-2-deoxy-alpha-D-glucopyranoside, RNA (5'-R(P*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Szychowski, J, Kondo, J, Zahr, O, Auclair, K, Westhof, E, Hanessian, S, Keillor, J.W. | | Deposit date: | 2011-05-20 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Inhibition of aminoglycoside-deactivating enzymes APH(3')-IIIa and AAC(6')-Ii by amphiphilic paromomycin O2''-ether analogues
Chemmedchem, 6, 2011
|
|
4PDQ
 
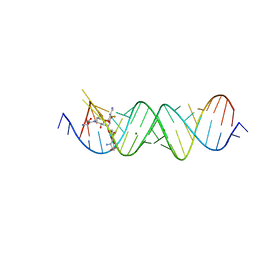 | | Crystal structure of the bacterial ribosomal decoding site in complex with 4'-deoxy-4'-fluoro neomycin analog | | Descriptor: | (2S)-4-amino-N-{(1R,2S,3R,4R,5S)-5-amino-3-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy }-4-[(2,6-diamino-2,4,6-trideoxy-4-fluoro-alpha-D-galactopyranosyl)oxy]-2-hydroxycyclohexyl}-2-hydroxybutanamide, MAGNESIUM ION, RNA (5'-*UP*UP*GP*CP*GP*UP*CP*AP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C-3') | | Authors: | Hanessian, S, Saavedra, O.M, Vilchis-Reyes, M.A, Maianti, J.P, Kanazawa, H, Dozzo, P, Feeney, L.A, Armstrong, E.S, Kondo, J. | | Deposit date: | 2014-04-21 | | Release date: | 2015-01-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis, broad spectrum antibacterial activity, and X-ray co-crystal structure of the decoding bacterial ribosomal A-site with 4'-deoxy-4'-fluoro neomycin analogs
Chem Sci, 5, 2014
|
|
5XZ1
 
 | |
5Z1I
 
 | | Crystal structure of the protozoal cytoplasmic ribosomal decoding site in complex with 6'-fluoro sisomicin | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(fluoromethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, RNA (5'-R(P*GP*CP*GP*UP*CP*GP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kanazawa, H, Hanessian, S, Kondo, J. | | Deposit date: | 2017-12-26 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Structure-Based Design of a Eukaryote-Selective Antiprotozoal Fluorinated Aminoglycoside.
ChemMedChem, 13, 2018
|
|
5Z71
 
 | |
5ZEJ
 
 | | Crystal structure of the bacterial A1408me1A-mutant ribosomal decoding site in complex with paromomycin | | Descriptor: | PAROMOMYCIN, RNA (5'-R(P*UP*GP*CP*GP*UP*CP*(1MA)P*CP*GP*UP*CP*GP*AP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kanazawa, H, Baba, F, Koganei, M, Kondo, J. | | Deposit date: | 2018-02-27 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A structural basis for the antibiotic resistance conferred by an N1-methylation of A1408 in 16S rRNA.
Nucleic Acids Res., 45, 2017
|
|
5ZEG
 
 | |
5ZEM
 
 | |
5ZEI
 
 | | Crystal structure of the bacterial A1408me1A-mutant ribosomal decoding site in complex with geneticin | | Descriptor: | GENETICIN, RNA (5'-R(P*GP*CP*GP*UP*CP*(1MA)P*CP*GP*UP*CP*GP*AP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3'), RNA (5'-R(P*UP*GP*CP*GP*UP*CP*(1MA)P*CP*GP*UP*CP*GP*AP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kanazawa, H, Baba, F, Koganei, M, Kondo, J. | | Deposit date: | 2018-02-27 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural basis for the antibiotic resistance conferred by an N1-methylation of A1408 in 16S rRNA.
Nucleic Acids Res., 45, 2017
|
|
5Z1H
 
 | | Crystal structure of the bacterial ribosomal decoding site in complex with 6'-fluoro sisomicin | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(fluoromethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kanazawa, H, Hanessian, S, Kondo, J. | | Deposit date: | 2017-12-26 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Design of a Eukaryote-Selective Antiprotozoal Fluorinated Aminoglycoside.
ChemMedChem, 13, 2018
|
|
6JBF
 
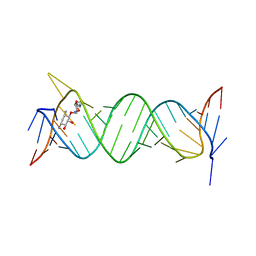 | | Crystal structure of the bacterial ribosomal decoding site in complex with 4'-deoxy-4'-fluoro neamine analog (axial 4'-F) | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2,6-diamino-2,4,6-trideoxy-4-fluoro-alpha-D-galactopyranoside, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kanazawa, H, Auffinger, P, Hanessian, S, Kondo, J. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the bacterial ribosomal decoding site in complex with 4'-deoxy-4'-fluoro neamine analog (axial 4'-F)
To Be Published
|
|
6JBG
 
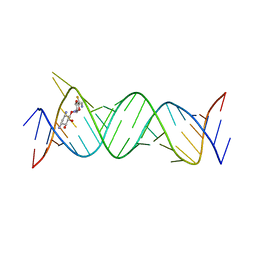 | | Crystal structure of the bacterial ribosomal decoding site in complex with 4'-deoxy-4'-fluoro neamine analog (equatorial 4'-F) | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2,6-diamino-2,4,6-trideoxy-4-fluoro-alpha-D-glucopyranoside, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3'), RNA (5'-R(P*UP*GP*CP*GP*UP*CP*AP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Kanazawa, H, Auffinger, P, Hanessian, S, Kondo, J. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the bacterial ribosomal decoding site in complex with 4'-deoxy-4'-fluoro neamine analog (axial 4'-F)
To Be Published
|
|
6IUE
 
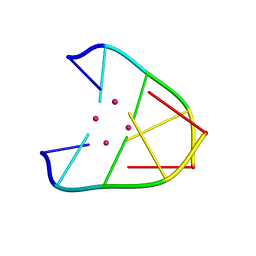 | | DNA helical wire containing Hg(II) | | Descriptor: | DNA (5'-D(*TP*TP*TP*GP*C)-3'), MERCURY (II) ION | | Authors: | Ono, A, Kanazawa, H, Ito, H, Goto, M, Nakamura, K, Saneyoshi, H, Kondo, J. | | Deposit date: | 2018-11-28 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | A Novel DNA Helical Wire Containing HgII-Mediated T:T and T:G Pairs.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
3WRU
 
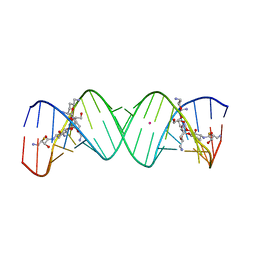 | | Crystal structure of the bacterial ribosomal decoding site in complex with synthetic aminoglycoside with F-HABA group | | Descriptor: | (2R,3R)-4-amino-N-[(1R,2S,3R,4R,5S)-5-amino-4-[(2,6-diamino-2,3,4,6-tetradeoxy-alpha-D-erythro-hexopyranosyl)oxy]-3-{[3-O-(2,6-diamino-2,3,4,6-tetradeoxy-beta-L-threo-hexopyranosyl)-beta-D-ribofuranosyl]oxy}-2-hydroxycyclohexyl]-3-fluoro-2-hydroxybutanamide, POTASSIUM ION, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*GP*CP*CP*GP*GP*CP*GP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Maianti, J.P, Kanazawa, H, Dozzo, P, Feeney, L.A, Armstrong, E.S, Kondo, J, Hanessian, S. | | Deposit date: | 2014-02-27 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Toxicity Modulation, Resistance Enzyme Evasion, and A-Site X-ray Structure of Broad-Spectrum Antibacterial Neomycin Analogs
Acs Chem.Biol., 9, 2014
|
|
1IXJ
 
 | | Crystal Structure of d(GCGAAAGCT) Containing Parallel-stranded Duplex with Homo Base Pairs and Anti-Parallel Duplex with Watson-Crick Base pairs | | Descriptor: | 5'-D(*GP*CP*GP*AP*AP*AP*GP*CP*T)-3', COBALT HEXAMMINE(III), MAGNESIUM ION | | Authors: | Sunami, T, Kondo, J, Kobuna, T, Hirao, I, Watanabe, K, Miura, K, Takenaka, A. | | Deposit date: | 2002-06-22 | | Release date: | 2002-12-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of d(GCGAAAGCT) Containing a Parallel-stranded Duplex with Homo Base Pairs and an Anti-Parallel Duplex with Watson-Crick Base pairs
Nucleic Acids Res., 30, 2002
|
|
