5F1P
 
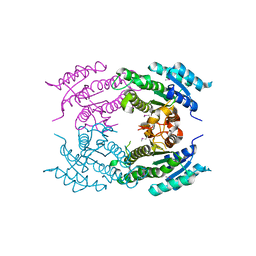 | | Crystal Structure of Dehydrogenase from Streptomyces platensis | | Descriptor: | PtmO8 | | Authors: | Kim, Y, Li, H, Endres, M, Babnigg, G, Rudolf, J, Ma, M, Chang, C.-Y, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-11-30 | | Release date: | 2015-12-30 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Crystal Structure of a Dehydrogenase, PtmO8, from Streptomyces platensis
To Be Published
|
|
4HL2
 
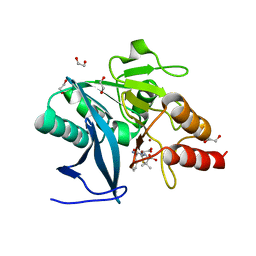 | | New Delhi Metallo-beta-Lactamase-1 1.05 A structure Complexed with Hydrolyzed Ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase NDM-1, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-10-15 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | New Delhi Metallo-beta-Lactamase-1 1.05 A structure Complexed with Hydrolyzed Ampicillin
To be Published
|
|
4HL1
 
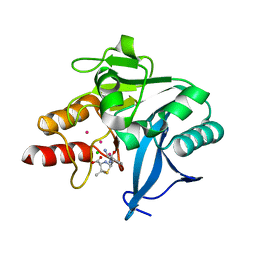 | | Crystal Structure of New Delhi Metallo-beta-Lactamase-1, Complexed with Cd and Ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Beta-lactamase NDM-1, CADMIUM ION, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-10-15 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of New Delhi Metallo-beta-Lactamase-1, Complexed with Cd and Ampicillin
To be Published
|
|
4HKY
 
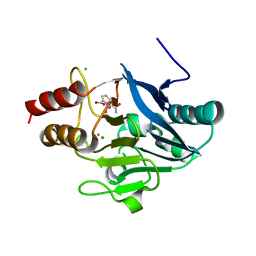 | | New Delhi Metallo-beta-Lactamase-1, Complexed with Cd and Faropenem | | Descriptor: | (2R)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-5-[(2R)-oxolan-2-yl]-2,3-dihydro-1,3-thiazole-4-carboxylic acid, (5R,6S)-6-(1-hydroxyethyl)-7-oxo-3-[(2R)-oxolan-2-yl]-4-thia-1-azabicyclo[3.2.0]hept-2-ene-2-carboxylic acid, Beta-lactamase NDM-1, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-10-15 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | New Delhi Metallo-beta-Lactamase-1, Complexed with Cd and Faropenem
To be Published
|
|
7F27
 
 | | Crystal structure of polyketide ketosynthase | | Descriptor: | 3-oxoacyl-(Acyl-carrier-protein) synthase | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2021-06-10 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.806 Å) | | Cite: | Structural basis of the complementary activity of two ketosynthases in aryl polyene biosynthesis.
Sci Rep, 11, 2021
|
|
4IZZ
 
 | | Crystal Structure of Fischerella Transcription Factor HetR complexed with 21mer DNA target | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*TP*GP*AP*GP*GP*GP*GP*TP*TP*AP*AP*AP*CP*CP*CP*CP*TP*CP*AP*C)-3'), SULFATE ION, ... | | Authors: | Kim, Y, Joachimiak, G, Gornicki, P, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structures of complexes comprised of Fischerella transcription factor HetR with Anabaena DNA targets.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4J00
 
 | | Crystal Structure of Fischerella Transcription Factor HetR complexed with 24mer DNA target | | Descriptor: | DNA (5'-D(*TP*GP*GP*TP*GP*AP*GP*GP*GP*GP*TP*TP*AP*AP*AP*CP*CP*CP*CP*TP*CP*AP*CP*C)-3'), MAGNESIUM ION, Transcription Factor HetR | | Authors: | Kim, Y, Joachimiak, G, Gornicki, P, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Structures of complexes comprised of Fischerella transcription factor HetR with Anabaena DNA targets.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7MTU
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus anthracis in the complex with IMP and the inhibitor P221 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, INOSINIC ACID, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-13 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from
Bacillus anthracis in the complex with IMP and the inhibitor P221
To Be Published
|
|
7MTX
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus anthracis in the complex with IMP and the inhibitor P176 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-{2-chloro-5-[({2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}carbamoyl)amino]phenyl}-beta-D-ribopyranosylamine, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-13 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from
Bacillus anthracis in the complex with IMP and the inhibitor P176
To Be Published
|
|
4IQE
 
 | | Crystal Structure of Carboxyvinyl-Carboxyphosphonate Phosphorylmutase from Bacillus anthracis str. Ames Ancestor | | Descriptor: | Carboxyvinyl-carboxyphosphonate phosphorylmutase | | Authors: | Kim, Y, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-11 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal Structure of Carboxyvinyl-Carboxyphosphonate Phosphorylmutase from Bacillus anthracis str. Ames Ancestor
To be Published
|
|
4IQD
 
 | | Crystal Structure of Carboxyvinyl-Carboxyphosphonate Phosphorylmutase from Bacillus anthracis | | Descriptor: | Carboxyvinyl-carboxyphosphonate phosphorylmutase, PYRUVIC ACID | | Authors: | Kim, Y, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-11 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Carboxyvinyl-Carboxyphosphonate Phosphorylmutase from Bacillus anthracis
To be Published
|
|
4IQI
 
 | | Crystal Structure of 7-cyano-7-deazaguanine Reductase, QueF from Vibrio cholerae O1 biovar El Tor complexed with cytosine | | Descriptor: | 6-AMINOPYRIMIDIN-2(1H)-ONE, CHLORIDE ION, NADPH-dependent 7-cyano-7-deazaguanine reductase, ... | | Authors: | Kim, Y, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-11 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of 7-cyano-7-deazaguanine Reductase, QueF from Vibrio cholerae O1 biovar El Tor complexed with cytosine
To be Published
|
|
4IQF
 
 | | Crystal Structure of Methyionyl-tRNA Formyltransferase from Bacillus anthracis | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, GLYCEROL, Methionyl-tRNA formyltransferase, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-11 | | Release date: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (2.378 Å) | | Cite: | Crystal Structure of Methyionyl-tRNA Formyltransferase from Bacillus anthracis
To be Published
|
|
4J01
 
 | | Crystal Structure of Fischerella Transcription Factor HetR complexed with 29mer DNA target | | Descriptor: | DNA (29-MER), SULFATE ION, Transcription Factor HetR | | Authors: | Kim, Y, Joachimiak, G, Gornicki, P, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.246 Å) | | Cite: | Structures of complexes comprised of Fischerella transcription factor HetR with Anabaena DNA targets.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HCI
 
 | | Uncharacterized Cupredoxin-like Domain Protein Cupredoxin_1 from Bacillus anthracis | | Descriptor: | Cupredoxin 1, GLYCEROL | | Authors: | Kim, Y, Maltseva, N, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-30 | | Release date: | 2012-10-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Uncharacterized Cupredoxin-like Domain Protein Cupredoxin_1 from Bacillus anthracis
To be Published
|
|
4HCJ
 
 | | Crystal Structure of ThiJ/PfpI Domain Protein from Brachyspira murdochii | | Descriptor: | CHLORIDE ION, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Kim, Y, Bigelow, L, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-09-30 | | Release date: | 2012-10-24 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystal Structure of ThiJ/PfpI Domain Protein from Brachyspira murdochii
To be Published
|
|
4I4J
 
 | | The structure of SgcE10, the ACP-polyene thioesterase involved in C-1027 biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, ACP-polyene thioesterase, D(-)-TARTARIC ACID, ... | | Authors: | Kim, Y, Bigelow, L, Bearden, J, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-27 | | Release date: | 2012-12-12 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (2.784 Å) | | Cite: | Crystal Structure of Thioesterase SgcE10 Supporting Common Polyene Intermediates in 9- and 10-Membered Enediyne Core Biosynthesis.
Acs Omega, 2, 2017
|
|
7N3C
 
 | | Crystal Structure of Human Fab S24-202 in the complex with the N-terminal Domain of Nucleocapsid protein from SARS CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, Nucleoprotein, ... | | Authors: | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-31 | | Release date: | 2021-07-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
7N3D
 
 | | Crystal Structure of Human Fab S24-1564 in the complex with the N-terminal Domain of Nucleocapsid protein from SARS CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Nucleoprotein, ... | | Authors: | Kim, Y, Maltseva, N, Tesar, C, Jedrzejczak, R, Dugan, H, Stamper, C, Wilson, P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-31 | | Release date: | 2021-07-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Epitopes recognition of SARS-CoV-2 nucleocapsid RNA binding domain by human monoclonal antibodies.
Iscience, 27, 2024
|
|
7K3M
 
 | | Crystal Structure of the Beta Lactamase Class D from Chitinophaga pinensis by Serial Crystallography | | Descriptor: | Beta-lactamase | | Authors: | Kim, Y, Sherrell, D.A, Johnson, J, Lavens, A, Maltseva, N, Endres, M, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-11 | | Release date: | 2020-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Beta Lactamase Class D from Chitinophaga pinensis by Serial Crystallography
To Be Published
|
|
4GYU
 
 | | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 A121F mutant from Klebsiella pneumoniae | | Descriptor: | Beta-lactamase NDM-1, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-09-05 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 A121F mutant from Klebsiella pneumoniae
To be Published, 2012
|
|
4H0D
 
 | | New Delhi Metallo-beta-Lactamase-1 Complexed with Mn from Klebsiella pneumoniae | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase NDM-1, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-09-07 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | New Delhi Metallo-beta-Lactamase-1 Complexed with Mn from Klebsiella pneumoniae
To be Published
|
|
7K1L
 
 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with Uridine-2',3'-Vanadate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Endres, M, Welk, L, Chang, C, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-07 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
7K1O
 
 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with Uridine-3',5'-Diphosphate | | Descriptor: | 1,2-ETHANEDIOL, 1-(3,5-di-O-phosphono-alpha-L-xylofuranosyl)pyrimidine-2,4(1H,3H)-dione, Uridylate-specific endoribonuclease | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Endres, M, Welk, L, Chang, C, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-09-08 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with Uridine-3',5'-Diphosphate
To Be Published
|
|
4HCG
 
 | | Uncharacterized Cupredoxin-like Domain Protein Cupredoxin_1 with Zinc bound from Bacillus anthracis | | Descriptor: | Cupredoxin 1, ZINC ION | | Authors: | Kim, Y, Maltseva, N, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-29 | | Release date: | 2012-10-17 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Uncharacterized Cupredoxin-like Domain Protein Cupredoxin_1 with Zinc bound from Bacillus anthracis
To be Published
|
|
