3M84
 
 | | Crystal Structure of Phosphoribosylaminoimidazole Synthetase from Francisella tularensis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Maltseva, N, Kim, Y, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-17 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal Structure of Phosphoribosylaminoimidazole Synthetase from
Francisella tularensis
To be Published
|
|
3CI6
 
 | | Crystal structure of the GAF domain from Acinetobacter phosphoenolpyruvate-protein phosphotransferase | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Cuff, M.E, Shackelford, G, Kim, Y, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-10 | | Release date: | 2008-05-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the GAF domain from Acinetobacter phosphoenolpyruvate-protein phosphotransferase.
TO BE PUBLISHED
|
|
1NNI
 
 | | Azobenzene Reductase from Bacillus subtilis | | Descriptor: | FLAVIN MONONUCLEOTIDE, hypothetical protein yhda | | Authors: | Cuff, M.E, Kim, Y, Maj, L, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-13 | | Release date: | 2003-07-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Azobenzene Reductase from Bacillus subtilis
To be Published, 2003
|
|
1R0O
 
 | | Crystal Structure of the Heterodimeric Ecdysone Receptor DNA-binding Complex | | Descriptor: | Ecdysone Response Element, Ecdysone receptor, Ultraspiracle protein, ... | | Authors: | Devarakonda, S, Harp, J.M, Kim, Y, Ozyhar, A, Rastinejad, F. | | Deposit date: | 2003-09-22 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure of the Heterodimeric Ecdysone Receptor DNA-binding Complex
Embo J., 22, 2003
|
|
1C98
 
 | | SOLUTION STRUCTURE OF NEUROMEDIN B | | Descriptor: | NEUROMEDIN B | | Authors: | Lee, S, Kim, Y. | | Deposit date: | 1999-08-01 | | Release date: | 1999-08-11 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of neuromedin B by (1)H nuclear magnetic resonance spectroscopy.
FEBS Lett., 460, 1999
|
|
3PEI
 
 | | Crystal Structure of Cytosol Aminopeptidase from Francisella tularensis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cytosol aminopeptidase, ... | | Authors: | Maltseva, N, Kim, Y, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-26 | | Release date: | 2010-12-01 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Cytosol Aminopeptidase from
Francisella tularensis
To be Published
|
|
3N8H
 
 | | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis | | Descriptor: | ACETIC ACID, ADENOSINE MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Maltseva, N, Kim, Y, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-28 | | Release date: | 2010-06-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis
To be Published, 2010
|
|
2Q6J
 
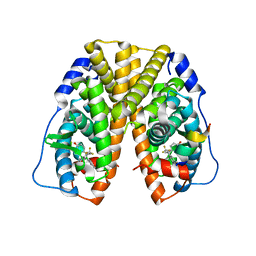 | | Crystal Structure of Estrogen Receptor alpha Complexed to a B-N Substituted Ligand | | Descriptor: | 4-[(DIMESITYLBORYL)(2,2,2-TRIFLUOROETHYL)AMINO]PHENOL, Estrogen receptor, GRIP peptide | | Authors: | Zhou, H, Nettles, K.W, Bruning, J.B, Kim, Y, Joachimiak, A, Sharma, S, Carlson, K.E, Stossi, F, Katzenellenbogen, B.S, Greene, G.L, Katzenellenbogen, J.A. | | Deposit date: | 2007-06-05 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Elemental isomerism: a boron-nitrogen surrogate for a carbon-carbon double bond increases the chemical diversity of estrogen receptor ligands
Chem.Biol., 14, 2007
|
|
1ODO
 
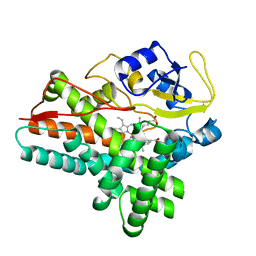 | | 1.85 A structure of CYP154A1 from Streptomyces coelicolor A3(2) | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, PUTATIVE CYTOCHROME P450 154A1 | | Authors: | Podust, L.M, Kim, Y, Arase, M, Bach, H, Sherman, D.H, Lamb, D.C, Kelly, S.L, Waterman, M.R. | | Deposit date: | 2003-02-19 | | Release date: | 2004-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Comparison of the 1.85 A Structure of Cyp154A1 from Streptomyces Coelicolor A3(2) with the Closely Related Cyp154C1 and Cyps from Antibiotic Biosynthetic Pathways.
Protein Sci., 13, 2004
|
|
1QPS
 
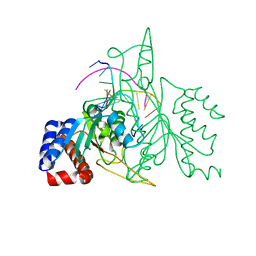 | | THE CRYSTAL STRUCTURE OF A POST-REACTIVE COGNATE DNA-ECO RI COMPLEX AT 2.50 A IN THE PRESENCE OF MN2+ ION | | Descriptor: | 5'-D(*AP*AP*TP*TP*CP*GP*CP*GP*)-3', 5'-D(*TP*CP*GP*CP*GP*)-3', ENDONUCLEASE ECORI, ... | | Authors: | Horvath, M, Choi, J, Kim, Y, Wilkosz, P, Rosenberg, J.M. | | Deposit date: | 1999-05-28 | | Release date: | 1999-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Integration of Recognition and Cleavage: X-Ray Structures of Pre- Transition State and Post-Reactive DNA-Eco RI Endonuclease Complexes
To be Published
|
|
5HW2
 
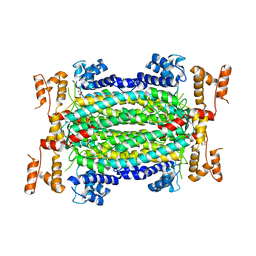 | | Crystal Structure of Adenylosuccinate Lyase from Francisella tularensis Complexed with fumaric acid | | Descriptor: | 1,2-ETHANEDIOL, Adenylosuccinate lyase, FUMARIC ACID, ... | | Authors: | Chang, C, Maltseva, N, Kim, Y, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-28 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Crystal Structure of Adenylosuccinate Lyase from Francisella tularensis Complexed with fumaric acid
To Be Published
|
|
5HG0
 
 | | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis complex with SAM | | Descriptor: | Pantothenate synthetase, S-ADENOSYLMETHIONINE | | Authors: | Chang, C, Maltseva, N, Kim, Y, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-07 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis complex with SAM
To Be Published
|
|
1R0N
 
 | | Crystal Structure of Heterodimeric Ecdsyone receptor DNA binding complex | | Descriptor: | Ecdsyone Response Element, Ecdysone Response Element, Ecdysone receptor, ... | | Authors: | Devarakonda, S, Harp, J.M, Kim, Y, Ozyhar, A, Rastinejad, F. | | Deposit date: | 2003-09-22 | | Release date: | 2003-10-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the heterodimeric Ecdysone Receptor DNA-binding complex
Embo J., 22, 2003
|
|
5HI6
 
 | | The high resolution structure of dihydrofolate reductase from Yersinia pestis complex with methotrexate as closed form | | Descriptor: | CALCIUM ION, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Chang, C, Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-11 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | The high resolution structure of dihydrofolate reductase from Yersinia pestis complex with methotrexate as closed form
To Be Published
|
|
5HD6
 
 | | High resolution structure of 3-hydroxydecanoyl-(acyl carrier protein) dehydratase from Yersinia pestis at 1.35 A | | Descriptor: | 3-hydroxydecanoyl-[acyl-carrier-protein] dehydratase, GLYCEROL | | Authors: | Chang, C, Maltseva, N, Kim, Y, Mulligan, R, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-04 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High resolution structure of 3-hydroxydecanoyl-(acyl carrier protein) dehydratase from Yersinia pestis at 1.35 A
To Be Published
|
|
5FAR
 
 | | Crystal structure of dihydroneopterin aldolase from Bacillus anthracis complex with 9-METHYLGUANINE | | Descriptor: | 7,8-dihydroneopterin aldolase, 9-METHYLGUANINE | | Authors: | Chang, C, Maltseva, N, Kim, Y, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-11 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of dihydroneopterin aldolase from Bacillus anthracis complex with 9-METHYLGUANINE
To Be Published
|
|
1QRH
 
 | | X-RAY STRUCTURE OF THE DNA-ECO RI ENDONUCLEASE COMPLEXES WITH AN R145K MUTATION AT 2.7 A | | Descriptor: | 5'-(TP*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G*)-3', ECO RI ENDONCULEASE | | Authors: | Choi, J, Kim, Y, Greene, P, Hager, P, Rosenberg, J.M. | | Deposit date: | 1999-06-14 | | Release date: | 1999-06-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray Structure of the DNA-Eco RI Endonuclease Complexes with the ED144 and RK145 Mutations
To be Published
|
|
3PNS
 
 | | Crystal Structure of Uridine Phosphorylase Complexed with Uracil from Vibrio cholerae O1 biovar El Tor | | Descriptor: | ACETIC ACID, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-11-19 | | Release date: | 2010-12-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Crystal Structure of Uridine Phosphorylase Complexed with Uracil from Vibrio cholerae O1 biovar El Tor
To be Published
|
|
3M49
 
 | | Crystal Structure of Transketolase Complexed with Thiamine Diphosphate from Bacillus anthracis | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Maltseva, N, Kim, Y, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-10 | | Release date: | 2010-04-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Transketolase Complexed with Thiamine Diphosphate from Bacillus anthracis
To be Published
|
|
3FH3
 
 | | Crystal structure of a putative ECF-type sigma factor negative effector from Bacillus anthracis str. Sterne | | Descriptor: | NICKEL (II) ION, putative ECF-type sigma factor negative effector | | Authors: | Nocek, B, Kim, Y, Joachimiak, G, Du, J, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-08 | | Release date: | 2009-01-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Crystal structure of a putative ECF-type sigma factor negative effector from Bacillus anthracis str. Sterne
To be Published
|
|
3FFY
 
 | | Putative tetrapyrrole (corrin/porphyrin) methyltransferase from Bacteroides fragilis. | | Descriptor: | Putative tetrapyrrole (Corrin/porphyrin) methylase, SULFATE ION | | Authors: | Osipiuk, J, Volkart, L, Cobb, G, Kim, Y, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-04 | | Release date: | 2008-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystal structure of putative tetrapyrrole (corrin/porphyrin) methyltransferase from Bacteroides fragilis.
To be Published
|
|
3O6V
 
 | | Crystal structure of Uridine Phosphorylase from Vibrio cholerae O1 biovar El Tor | | Descriptor: | FORMIC ACID, GLYCEROL, Uridine phosphorylase | | Authors: | Maltseva, N, Kim, Y, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-29 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Crystal structure of Uridine Phosphorylase from Vibrio cholerae O1 biovar El Tor
To be Published
|
|
3Q1H
 
 | | Crystal Structure of Dihydrofolate Reductase from Yersinia pestis | | Descriptor: | Dihydrofolate reductase, SULFATE ION | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-17 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Crystal Structure of Dihydrofolate Reductase from Yersinia pestis
To be Published
|
|
2HLY
 
 | | The crystal structure of genomics APC5867 | | Descriptor: | Hypothetical protein Atu2299 | | Authors: | Dong, A, Xu, X, Zheng, H, Kim, Y, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-07-10 | | Release date: | 2006-07-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of genomics APC5867
TO BE PUBLISHED
|
|
3QTT
 
 | | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis Complexed with Beta-gamma ATP and Beta-alanine | | Descriptor: | BETA-ALANINE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Maltseva, N, Kim, Y, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-02-23 | | Release date: | 2011-03-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis Complexed with Beta-gamma ATP and Beta-alanine.
To be Published
|
|
