1R4V
 
 | | 1.9A crystal structure of protein AQ328 from Aquifex aeolicus | | Descriptor: | CACODYLATE ION, Hypothetical protein AQ_328, ZINC ION | | Authors: | Qiu, Y, Tereshko, V, Kim, Y, Zhang, R, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-10-08 | | Release date: | 2004-03-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of Aq_328 from the hyperthermophilic bacteria Aquifex aeolicus shows an ancestral histone fold.
Proteins, 62, 2006
|
|
2LNC
 
 | | Solution NMR structure of Norwalk virus protease | | Descriptor: | 3C-like protease | | Authors: | Takahashi, D, Hiromasa, Y, Kim, Y, Anbanandam, A, Chang, K, Prakash, O. | | Deposit date: | 2011-12-22 | | Release date: | 2012-12-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamics characterization of norovirus protease.
Protein Sci., 22, 2013
|
|
3IGJ
 
 | | Crystal Structure of Maltose O-acetyltransferase Complexed with Acetyl Coenzyme A from Bacillus anthracis | | Descriptor: | ACETYL COENZYME *A, FORMIC ACID, GLYCEROL, ... | | Authors: | Maltseva, N, Kim, Y, Papazisi, L, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-27 | | Release date: | 2009-08-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Maltose O-acetyltransferase Complexed with Acetyl Coenzyme A from Bacillus anthracis
To be Published
|
|
2N50
 
 | | Novel Structural Components Contribute to the High Thermal Stability of Acyl Carrier Protein from Enterococcus faecalis | | Descriptor: | Acyl carrier protein | | Authors: | Park, Y, Jung, M, Song, H, Jeong, K, Kim, Y. | | Deposit date: | 2015-07-02 | | Release date: | 2015-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Novel Structural Components Contribute to the High Thermal Stability of Acyl Carrier Protein from Enterococcus faecalis.
J. Biol. Chem., 291, 2016
|
|
3IQT
 
 | | Structure of the HPT domain of Sensor protein barA from Escherichia coli CFT073. | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Signal transduction histidine-protein kinase barA | | Authors: | Cuff, M.E, Rakowski, E, Kim, Y, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-08-20 | | Release date: | 2009-09-22 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the HPT domain of Sensor protein barA from Escherichia coli CFT073.
TO BE PUBLISHED
|
|
3IYD
 
 | | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator, DNA (98-MER), ... | | Authors: | Hudson, B.P, Quispe, J, Lara, S, Kim, Y, Berman, H, Arnold, E, Ebright, R.H, Lawson, C.L. | | Deposit date: | 2009-08-01 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (19.799999 Å) | | Cite: | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8GHY
 
 | | Crystal Structure of the E154D mutant CelD Cellulase from the Anaerobic Fungus Piromyces finnis in the complex with cellotriose. | | Descriptor: | Cellulase CelD, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Dementieve, A, Kim, Y, Jedrzejczak, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and enzymatic characterization of CelD endoglucanase from the anaerobic fungus Piromyces finnis.
Appl.Microbiol.Biotechnol., 107, 2023
|
|
8GHX
 
 | | Crystal Structure of CelD Cellulase from the Anaerobic Fungus Piromyces finnis | | Descriptor: | 1,2-ETHANEDIOL, Cellulase CelD | | Authors: | Dementieve, A, Kim, Y, Jedrzejczak, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure and enzymatic characterization of CelD endoglucanase from the anaerobic fungus Piromyces finnis.
Appl.Microbiol.Biotechnol., 107, 2023
|
|
2BPO
 
 | | Crystal structure of the yeast CPR triple mutant: D74G, Y75F, K78A. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Yermalitskaya, L.V, Kim, Y, Waterman, M.R, Podust, L.M. | | Deposit date: | 2005-04-21 | | Release date: | 2006-07-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Yeast Cpr Triple Mutant: D74G, Y75F, K78A.
To be Published
|
|
8GMH
 
 | | Crystal Structure of the ternary complex of TelA-LXG, LapA3, and LapA4 | | Descriptor: | 1,2-ETHANEDIOL, LXG domain-containing protein, LapA3, ... | | Authors: | Klein, T.A, Shah, P.Y, Gkragkopoulou, P, Grebenc, D.W, Kim, Y, Whitney, J.C. | | Deposit date: | 2023-03-25 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a tripartite protein complex that targets toxins to the type VII secretion system.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6MN5
 
 | | Crystal structure of aminoglycoside acetyltransferase AAC(3)-IVa, H154A mutant, in complex with gentamicin C1A | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 1,2-ETHANEDIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Kim, Y, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
2CI0
 
 | | High throughput screening and x-ray crystallography assisted evaluation of small molecule scaffolds for CYP51 inhibitors | | Descriptor: | (2R)-2-PHENYL-N-PYRIDIN-4-YLBUTANAMIDE, CYTOCHROME P450 51, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Podust, L.M, Kim, Y, Yermalitskaya, L.V, Von Kries, J.P, Waterman, M.R. | | Deposit date: | 2006-03-16 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Small Molecule Scaffolds for Cyp51 Inhibitors Identified by High Throughput Screening and Defined by X-Ray Crystallography
Antimicrob.Agents Chemother., 51, 2007
|
|
2C7X
 
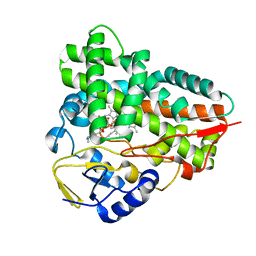 | | Crystal structure of narbomycin-bound cytochrome P450 PikC (CYP107L1) | | Descriptor: | CYTOCHROME P450 MONOOXYGENASE, NARBOMYCIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sherman, D.H, Li, S, Yermalitskaya, L.V, Kim, Y, Smith, J.A, Waterman, M.R, Podust, L.M. | | Deposit date: | 2005-11-29 | | Release date: | 2006-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Structural Basis for Substrate Anchoring, Active Site Selectivity, and Product Formation by P450 Pikc from Streptomyces Venezuelae.
J.Biol.Chem., 281, 2006
|
|
2CA0
 
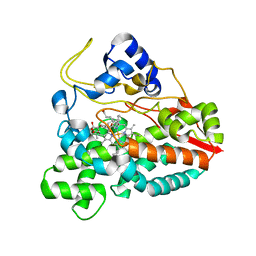 | | Crystal structure of YC-17-bound cytochrome P450 PikC (CYP107L1) | | Descriptor: | 4-{[4-(DIMETHYLAMINO)-3-HYDROXY-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL]OXY}-12-ETHYL-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, CYTOCHROME P450 MONOOXYGENASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yermalitskaya, L.I, Kim, Y, Sherman, D.H, Waterman, M.R, Podust, L.M. | | Deposit date: | 2005-12-15 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of Yc-17-Bound Cytochrome P450 Pikc (Cyp107L1)
To be Published
|
|
2CD8
 
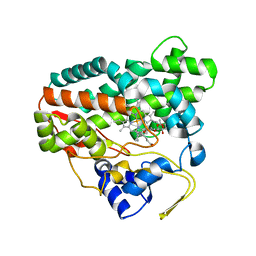 | | Crystal structure of YC-17-bound cytochrome P450 PikC (CYP107L1) | | Descriptor: | 4-{[4-(DIMETHYLAMINO)-3-HYDROXY-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL]OXY}-12-ETHYL-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, CYTOCHROME P450 MONOOXYGENASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yermalitskaya, L.I, Kim, Y, Sherman, D.H, Waterman, M.R, Podust, L.M. | | Deposit date: | 2006-01-20 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structural Basis for Substrate Anchoring, Active Site Selectivity, and Product Formation by P450 Pikc from Streptomyces Venezuelae.
J.Biol.Chem., 281, 2006
|
|
2C6H
 
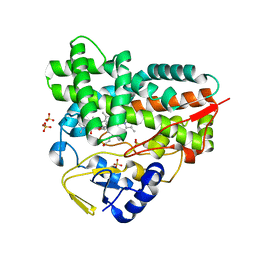 | | Crystal structure of YC-17-bound cytochrome P450 PikC (CYP107L1) | | Descriptor: | 4-{[4-(DIMETHYLAMINO)-3-HYDROXY-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL]OXY}-12-ETHYL-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, CYTOCHROME P450 MONOOXYGENASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Sherman, D.H, Li, S, Yermalitskaya, L.V, Kim, Y, Smith, J.A, Waterman, M.R, Podust, L.M. | | Deposit date: | 2005-11-09 | | Release date: | 2006-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Structural Basis for Substrate Anchoring, Active Site Selectivity, and Product Formation by P450 Pikc from Streptomyces Venezuelae.
J.Biol.Chem., 281, 2006
|
|
2BF4
 
 | | A second FMN-binding site in yeast NADPH-cytochrome P450 reductase suggests a novel mechanism of electron transfer by diflavin reductases. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Podust, L.M, Lepesheva, G.I, Kim, Y, Yermalitskaya, L.V, Yermalitsky, V.N, Lamb, D.C, Kelly, S.L, Waterman, M.R. | | Deposit date: | 2004-12-03 | | Release date: | 2006-01-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Second Fmn-Binding Site in Yeast Nadph-Cytochrome P450 Reductase Suggests a Mechanism of Electron Transfer by Diflavin Reductases.
Structure, 14, 2006
|
|
2BVJ
 
 | | Ligand-free structure of cytochrome P450 PikC (CYP107L1) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BETA-MERCAPTOETHANOL, CYTOCHROME P450 MONOOXYGENASE, ... | | Authors: | Sherman, D.H, Li, S, Yermalitskaya, L.V, Kim, Y, Smith, J.A, Waterman, M.R, Podust, L.M. | | Deposit date: | 2005-06-28 | | Release date: | 2006-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structural Basis for Substrate Anchoring, Active Site Selectivity, and Product Formation by P450 Pikc from Streptomyces Venezuelae.
J.Biol.Chem., 281, 2006
|
|
3HYL
 
 | | Crystal Structure of Transketolase from Bacillus anthracis | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Kwon, K, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-06-22 | | Release date: | 2009-06-30 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal Structure of Transketolase from Bacillus anthracis
To be Published
|
|
3HLV
 
 | | Crystal structure of human Estrogen Receptor Alpha Ligand-Binding Domain in complex with a Glucocorticoid Receptor Interacting Protein 1 Nr Box II Peptide and 16-alpha-hydroxy-estrone ((8S,9R,13S,14R,16R)-3,16-dihydroxy-13-methyl-7,8,9,11,12,14,15, 16-octahydro-6H-cyclopenta[a]phenanthren-17-one | | Descriptor: | (9beta,13alpha,16beta)-3,16-dihydroxyestra-1,3,5(10)-trien-17-one, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Rajan, S.S, Kim, Y, Vanek, K, Joachimiak, A, Greene, G.L. | | Deposit date: | 2009-05-28 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human alphaER-LBD in complex with GRIP peptide and 16alpha-hydroxyestrone
To be Published
|
|
3HM1
 
 | | Crystal structure of human Estrogen Receptor Alpha Ligand-Binding Domain in complex with a Glucocorticoid Receptor Interacting Protein 1 Nr Box II Peptide and estrone ((8R,9S,13S,14S)-3-hydroxy-13-methyl-7,8,9,11,12,14,15,16-octahydro-6H-cyclopenta[a]phenanthren-17-one) | | Descriptor: | (9beta,13alpha)-3-hydroxyestra-1,3,5(10)-trien-17-one, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Rajan, S.S, Kim, Y, Vanek, K, Liwanag, M, Joachimiak, A, Greene, G.L. | | Deposit date: | 2009-05-28 | | Release date: | 2010-06-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of human Estrogen Receptor Alpha Ligand-Binding Domain in complex with a Glucocorticoid Receptor Interacting Protein 1 Nr Box II Peptide and estrone ((8R,9S,13S,14S)-3-hydroxy-13-methyl-7,8,9,11,12,14,15, 16-octahydro-6H-cyclopenta[a]phenanthren-17-one)
To be Published
|
|
3L03
 
 | | Crystal Structure of human Estrogen Receptor alpha Ligand-Binding Domain in complex with a Glucocorticoid Receptor Interacting Protein 1 Nr Box II peptide and Estetrol (Estra-1,3,5(10)-triene-3,15 alpha,16alpha,17beta-tetrol) | | Descriptor: | (14beta,15alpha,16alpha,17alpha)-estra-1,3,5(10)-triene-3,15,16,17-tetrol, CHLORIDE ION, Estrogen receptor, ... | | Authors: | Rajan, S.S, Kim, Y, Vanek, K, Joachimiak, A, Greene, G.L. | | Deposit date: | 2009-12-09 | | Release date: | 2010-12-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Crystal Structure of human Estrogen Receptor alpha Ligand-Binding Domain
in complex with a Glucocorticoid Receptor Interacting Protein 1 Nr Box II
peptide and Estra-1,3,5(10)-triene-3,15 alpha,16alpha,17beta-tetrol
To be Published
|
|
2CIB
 
 | | High throughput screening and x-ray crystallography assisted evaluation of small molecule scaffolds for CYP51 inhibitors | | Descriptor: | (2S)-2-[(2,1,3-BENZOTHIADIAZOL-4-YLSULFONYL)AMINO]-2-PHENYL-N-PYRIDIN-4-YLACETAMIDE, CYTOCHROME P450 51, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Podust, L.M, Kim, Y, Yermalitskaya, L.V, Von Kries, J.P, Waterman, M.R. | | Deposit date: | 2006-03-17 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Small Molecule Scaffolds for Cyp51 Inhibitors Identified by High Throughput Screening and Defined by X-Ray Crystallography
Antimicrob.Agents Chemother., 51, 2007
|
|
2QH6
 
 | | Crystal Structure of the Estrogen Receptor Alpha Ligand Binding Domain Complexed with an Oxabicyclic diarylethylene Compound | | Descriptor: | DIETHYL (1R,2S,3R,4S)-5,6-BIS(4-HYDROXYPHENYL)-7-OXABICYCLO[2.2.1]HEPT-5-ENE-2,3-DICARBOXYLATE, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nettles, K.W, Bruning, J.B, Nowak, J, Sharma, S.K, Hahm, J.B, Shi, Y, Kulp, K, Hochberg, R.B, Zhou, H, Katzenellenbogen, J.A, Katzenellenbogen, B.S, Kim, Y, Joachmiak, A, Greene, G.L. | | Deposit date: | 2007-06-29 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | NFkappaB selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses
Nat.Chem.Biol., 4, 2008
|
|
2QXM
 
 | | Crystal Structure of the Estrogen Receptor Alpha Ligand Binding Domain Complexed to Burned Meat Compound PhIP | | Descriptor: | 2-AMINO-1-METHYL-6-PHENYLIMIDAZO[4,5-B]PYRIDINE, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nettles, K.W, Bruning, J.B, Gil, G, Nowak, J, Sharma, S.K, Hahm, J.B, Shi, Y, Kulp, K, Hochberg, R.B, Zhou, H, Katzenellenbogen, J.A, Katzenellenbogen, B.S, Kim, Y, Joachmiak, A, Greene, G.L. | | Deposit date: | 2007-08-12 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NFkappaB selectivity of estrogen receptor ligands revealed by comparative crystallographic analyses
Nat.Chem.Biol., 4, 2008
|
|
