4LFU
 
 | | Crystal structure of Escherichia coli SdiA in the space group C2 | | Descriptor: | CHLORIDE ION, Regulatory protein SdiA, TETRAETHYLENE GLYCOL | | Authors: | Kim, T, Duong, T, Wu, C.A, Choi, J, Lan, N, Kang, S.W, Lokanath, N.K, Shin, D, Hwang, H.Y, Kim, K.K. | | Deposit date: | 2013-06-27 | | Release date: | 2014-03-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural insights into the molecular mechanism of Escherichia coli SdiA, a quorum-sensing receptor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LGW
 
 | | Crystal structure of Escherichia coli SdiA in the space group P6522 | | Descriptor: | GLYCEROL, Regulatory protein SdiA | | Authors: | Kim, T, Duong, T, Wu, C.A, Choi, J, Lan, N, Kang, S.W, Lokanath, N.K, Shin, D, Hwang, H.Y, Kim, K.K. | | Deposit date: | 2013-06-28 | | Release date: | 2014-03-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the molecular mechanism of Escherichia coli SdiA, a quorum-sensing receptor
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5IS1
 
 | |
4N06
 
 | |
7W8E
 
 | |
7W8H
 
 | |
3FAU
 
 | | Crystal Structure of human small-MutS related domain | | Descriptor: | NEDD4-binding protein 2 | | Authors: | Kim, T.G, Kwon, T.H, Ryu, E.K, Min, K, Heo, S.-D, Song, K.M, Jun, W.J, Jung, E. | | Deposit date: | 2008-11-18 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Strcutral Dynamincs of the Endonuclease Small-MutS Related Domains of BCL3 binding protein
To be Published
|
|
6JYI
 
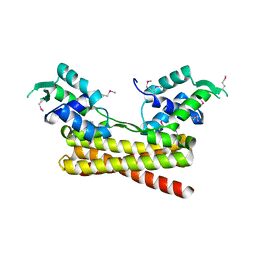 | | Crystal structure of the PadR-like transcriptional regulator BC1756 from Bacillus cereus | | Descriptor: | Transcriptional repressor PadR | | Authors: | Kim, T.H, Park, S.C, Lee, K.C, Song, W.S, Yoon, S.I. | | Deposit date: | 2019-04-26 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural and DNA-binding studies of the PadR-like transcriptional regulator BC1756 from Bacillus cereus.
Biochem.Biophys.Res.Commun., 515, 2019
|
|
8YBK
 
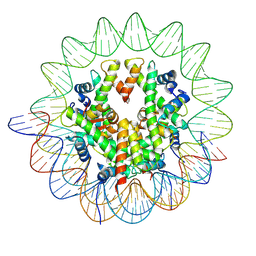 | | Cryo-EM structure of the human nucleosome containing the H3.1 E97K mutant | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kimura, T, Hirai, S, Kujirai, T, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2024-02-14 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Cryo-EM structure and biochemical analyses of the nucleosome containing the cancer-associated histone H3 mutation E97K.
Genes Cells, 29, 2024
|
|
8YBJ
 
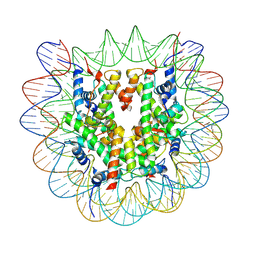 | | Cryo-EM structure of human nucleosome core particle composed of the Widom 601 DNA sequence | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kimura, T, Hirai, S, Kujirai, T, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2024-02-14 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Cryo-EM structure and biochemical analyses of the nucleosome containing the cancer-associated histone H3 mutation E97K.
Genes Cells, 29, 2024
|
|
6A94
 
 | | Crystal structure of 5-HT2AR in complex with zotepine | | Descriptor: | 2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(3-chloranylbenzo[b][1]benzothiepin-5-yl)oxy-N,N-dimethyl-ethanamine, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Kimura, T.K, Asada, H, Inoue, A, Kadji, F.M.N, Im, D, Mori, C, Arakawa, T, Hirata, K, Nomura, Y, Nomura, N, Aoki, J, Iwata, S, Shimamura, T. | | Deposit date: | 2018-07-11 | | Release date: | 2019-02-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the 5-HT2Areceptor in complex with the antipsychotics risperidone and zotepine.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6A93
 
 | | Crystal structure of 5-HT2AR in complex with risperidone | | Descriptor: | 3-[2-[4-(6-fluoranyl-1,2-benzoxazol-3-yl)piperidin-1-yl]ethyl]-2-methyl-6,7,8,9-tetrahydropyrido[1,2-a]pyrimidin-4-one, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, CHOLESTEROL, ... | | Authors: | Kimura, T.K, Asada, H, Inoue, A, Kadji, F.M.N, Im, D, Mori, C, Arakawa, T, Hirata, K, Nomura, Y, Nomura, N, Aoki, J, Iwata, S, Shimamura, T. | | Deposit date: | 2018-07-11 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the 5-HT2Areceptor in complex with the antipsychotics risperidone and zotepine.
Nat.Struct.Mol.Biol., 26, 2019
|
|
3A8W
 
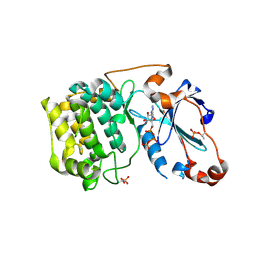 | | Crystal Structure of PKCiota kinase domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Protein kinase C iota type, SULFATE ION | | Authors: | Takimura, T, Kamata, K. | | Deposit date: | 2009-10-11 | | Release date: | 2010-05-05 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the PKC-iota kinase domain in its ATP-bound and apo forms reveal defined structures of residues 533-551 in the C-terminal tail and their roles in ATP binding
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3A8X
 
 | | Crystal Structure of PKCiota kinase domain | | Descriptor: | Protein kinase C iota type, SULFATE ION | | Authors: | Takimura, T, Kamata, K. | | Deposit date: | 2009-10-11 | | Release date: | 2010-05-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the PKC-iota kinase domain in its ATP-bound and apo forms reveal defined structures of residues 533-551 in the C-terminal tail and their roles in ATP binding
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2Z1Q
 
 | |
4N7Z
 
 | | Crystal structure of human Plk4 cryptic polo box (CPB) in complex with a Cep192 N-terminal fragment | | Descriptor: | Centrosomal protein of 192 kDa, Serine/threonine-protein kinase PLK4 | | Authors: | Park, S.-Y, Park, J.-E, Tian, L, Kim, T.-S, Yang, W, Lee, K.S. | | Deposit date: | 2013-10-16 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis for unidirectional scaffold switching of human Plk4 in centriole biogenesis.
Nat.Struct.Mol.Biol., 21, 2014
|
|
8DOA
 
 | |
4XSJ
 
 | | Crystal structure of the N-terminal domain of the human mitochondrial calcium uniporter fused with T4 lysozyme | | Descriptor: | Lysozyme,Calcium uniporter protein, mitochondrial, SULFATE ION | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-01-22 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
4XTB
 
 | | Crystal structure of the N-terminal domain of the human mitochondrial calcium uniporter | | Descriptor: | Calcium uniporter protein, mitochondrial, TETRAETHYLENE GLYCOL | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-01-23 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
4MM2
 
 | | Crystal structure of yeast primase catalytic subunit | | Descriptor: | CADMIUM ION, CITRIC ACID, DNA primase small subunit | | Authors: | Park, K.R, An, J.Y, Lee, Y, Youn, H.S, Lee, J.G, Kang, J.Y, Kim, T.G, Lim, J.J, Eom, S.H, Wang, J. | | Deposit date: | 2013-09-07 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of yeast primase catalytic subunit
To be Published
|
|
5JNM
 
 | | Crystal structure of MtlD from Staphylococcus aureus at 1.7-Angstrom resolution | | Descriptor: | Mannitol-1-phosphate 5-dehydrogenase, SULFATE ION | | Authors: | Ta, H.M, Nguyen, T, Kim, T, Kim, K.K. | | Deposit date: | 2016-04-30 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Targeting Mannitol Metabolism as an Alternative Antimicrobial Strategy Based on the Structure-Function Study of Mannitol-1-Phosphate Dehydrogenase in Staphylococcus aureus.
Mbio, 10, 2019
|
|
5BZ6
 
 | | Crystal structure of the N-terminal domain single mutant (S92A) of the human mitochondrial calcium uniporter fused with T4 lysozyme | | Descriptor: | Lysozyme,Calcium uniporter protein, mitochondrial, SULFATE ION | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-06-11 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
5HOE
 
 | | Crystal structrue of Est24, a carbohydrate acetylesterase from Sinorhizobium meliloti | | Descriptor: | HEXAETHYLENE GLYCOL, Hydrolase, PHOSPHATE ION | | Authors: | Oh, C, Ryu, B.H, An, D.R, Nguyen, D.D, Yoo, W, Kim, T, Ngo, D.T, Kim, H.S, Park, J.S, Kim, K.K, Kim, T.D. | | Deposit date: | 2016-01-19 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Biochemical Characterization of an Octameric Carbohydrate Acetylesterase from Sinorhizobium meliloti.
Febs Lett., 590, 2016
|
|
5SWN
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Asp110Asn/Fluoroacetate - Cocrystallized | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase, fluoroacetic acid | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-08-08 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
5T4T
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Asp110Asn - Apo No Halide | | Descriptor: | ACETATE ION, Fluoroacetate dehalogenase | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-08-30 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
