8CV0
 
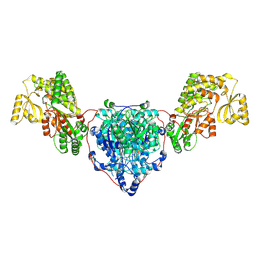 | | KS-AT domains of mycobacterial Pks13 with outward AT conformation | | Descriptor: | Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CV1
 
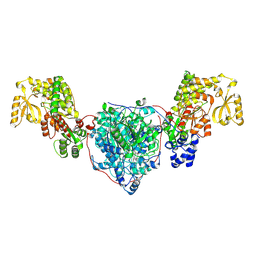 | | ACP1-KS-AT domains of mycobacterial Pks13 | | Descriptor: | Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8CUZ
 
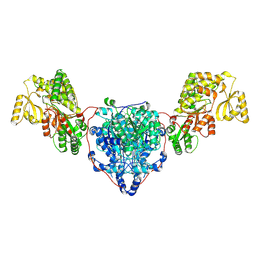 | | KS-AT domains of mycobacterial Pks13 with inward AT conformation | | Descriptor: | Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5Y9D
 
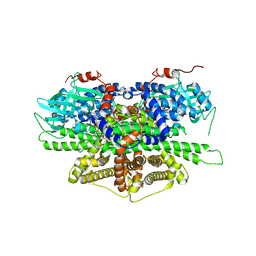 | | Crystal structure of acyl-coA oxidase1 from Yarrowia lipolytica | | Descriptor: | Acyl-coenzyme A oxidase 1, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kim, S, Kim, K.-J. | | Deposit date: | 2017-08-24 | | Release date: | 2018-01-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into the substrate specificity of acyl-CoA oxidase1 from Yarrowia lipolytica for short-chain dicarboxylyl-CoAs.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
3GEB
 
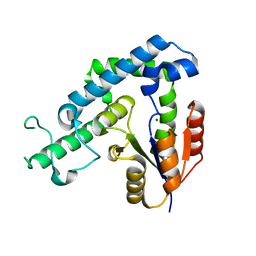 | | Crystal Structure of edeya2 | | Descriptor: | Eyes absent homolog 2, MAGNESIUM ION | | Authors: | Kim, S.J. | | Deposit date: | 2009-02-25 | | Release date: | 2009-04-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of edeya2
To be Published
|
|
2F6L
 
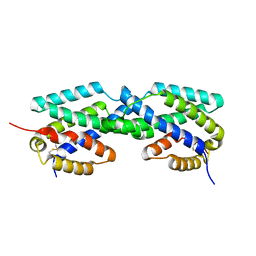 | | X-ray structure of Chorismate Mutase from Mycobacterium Tuberculosis | | Descriptor: | Chorismate mutase | | Authors: | Kim, S.K, Howard, A.J, Gilliland, G.L, Reddy, P.T, Ladner, J.E. | | Deposit date: | 2005-11-29 | | Release date: | 2005-12-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural characterization of the secreted chorismate mutase (Rv1885c) from Mycobacterium tuberculosis H37Rv: an *AroQ enzyme not regulated by the aromatic amino acids.
J.Bacteriol., 188, 2006
|
|
7VID
 
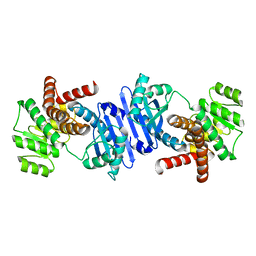 | |
1JJ4
 
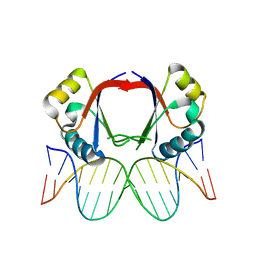 | | Human papillomavirus type 18 E2 DNA-binding domain bound to its DNA target | | Descriptor: | E2 binding site DNA, Regulatory protein E2 | | Authors: | Kim, S.-S, Tam, J.K, Wang, A.F, Hegde, R.S. | | Deposit date: | 2001-07-03 | | Release date: | 2001-07-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis of DNA target discrimination by papillomavirus E2 proteins.
J.Biol.Chem., 275, 2000
|
|
2I1V
 
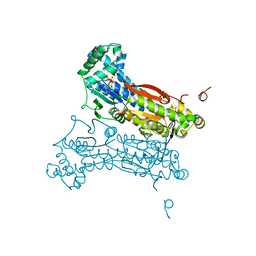 | | Crystal structure of PFKFB3 in complex with ADP and Fructose-2,6-bisphosphate | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3, ... | | Authors: | Kim, S.G, El-Maghrabi, M.R, Lee, Y.H. | | Deposit date: | 2006-08-15 | | Release date: | 2007-07-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Direct Substrate-Substrate Interaction Found in the Kinase Domain of the Bifunctional Enzyme, 6-Phosphofructo-2-kinase/Fructose-2,6-bisphosphatase
J.Mol.Biol., 370, 2007
|
|
1R1A
 
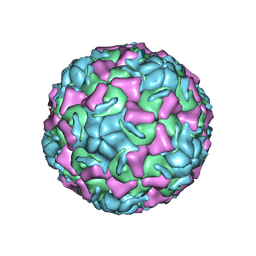 | | CRYSTAL STRUCTURE OF HUMAN RHINOVIRUS SEROTYPE 1A (HRV1A) | | Descriptor: | HUMAN RHINOVIRUS 1A COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 1A COAT PROTEIN (SUBUNIT VP2), HUMAN RHINOVIRUS 1A COAT PROTEIN (SUBUNIT VP3), ... | | Authors: | Kim, S, Rossmann, M.G. | | Deposit date: | 1989-03-15 | | Release date: | 1990-07-15 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of human rhinovirus serotype 1A (HRV1A).
J.Mol.Biol., 210, 1989
|
|
3OTP
 
 | |
3ACP
 
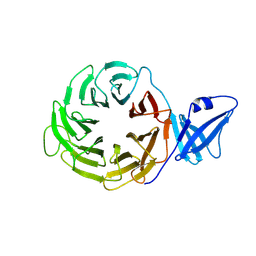 | | Crystal Structure of Yeast Rpn14, a Chaperone of the 19S Regulatory Particle of the Proteasome | | Descriptor: | WD repeat-containing protein YGL004C | | Authors: | Kim, S, Saeki, Y, Suzuki, A, Takagi, K, Fukunaga, K, Yamane, T, Kato, K, Tanaka, K, Mizushima, T. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of yeast Rpn14, a chaperone of the 19S regulatory particle of the proteasome
J.Biol.Chem., 285, 2010
|
|
3VL1
 
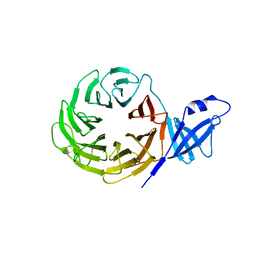 | | Crystal structure of yeast Rpn14 | | Descriptor: | 26S proteasome regulatory subunit RPN14 | | Authors: | Kim, S, Nishide, A, Saeki, Y, Takagi, K, Tanaka, K, Kato, K, Mizushima, T. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New crystal structure of the proteasome-dedicated chaperone Rpn14 at 1.6 A resolution
Acta Crystallogr.,Sect.F, 68, 2012
|
|
1I7F
 
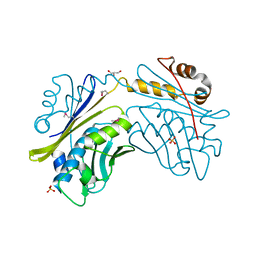 | | CRYSTAL STRUCTURE OF THE HSP33 DOMAIN WITH CONSTITUTIVE CHAPERONE ACTIVITY | | Descriptor: | GLYCEROL, HEAT SHOCK PROTEIN 33, SULFATE ION | | Authors: | Kim, S.-J, Jeong, D.-G, Chi, S.-W, Lee, J.-S, Ryu, S.-E. | | Deposit date: | 2001-03-09 | | Release date: | 2001-05-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of proteolytic fragments of the redox-sensitive Hsp33 with constitutive chaperone activity
Nat.Struct.Biol., 8, 2001
|
|
6KIA
 
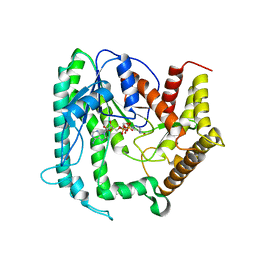 | |
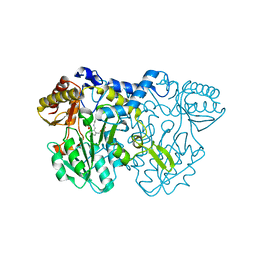
4LW4
 
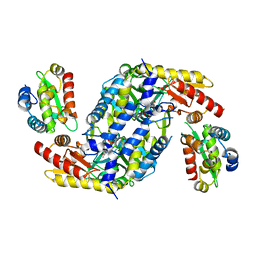 | |
4LW2
 
 | |
7UK4
 
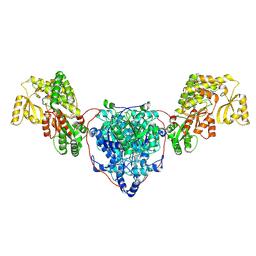 | | KS-AT di-domain of mycobacterial Pks13 with endogenous KS ligand bound | | Descriptor: | Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-03-31 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (1.94 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2MLP
 
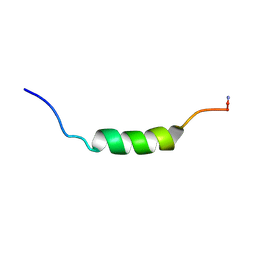 | | MICROCIN LEADER PEPTIDE FROM E. COLI, NMR, 25 STRUCTURES | | Descriptor: | MCBA PROPEPTIDE | | Authors: | Kim, S, Sinha Roy, R, Walsh, C.T, Baleja, J.D. | | Deposit date: | 1998-01-21 | | Release date: | 1998-07-22 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Role of the microcin B17 propeptide in substrate recognition: solution structure and mutational analysis of McbA1-26.
Chem.Biol., 5, 1998
|
|
1F9F
 
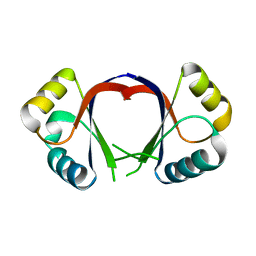 | | CRYSTAL STRUCTURE OF THE HPV-18 E2 DNA-BINDING DOMAIN | | Descriptor: | Regulatory protein E2, SULFATE ION | | Authors: | Kim, S.S, Tam, J, Wang, A.F, Hegde, R. | | Deposit date: | 2000-07-10 | | Release date: | 2000-11-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis of DNA target discrimination by papillomavirus E2 proteins.
J.Biol.Chem., 275, 2000
|
|
1RYU
 
 | | Solution Structure of the SWI1 ARID | | Descriptor: | SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin subfamily F member 1 | | Authors: | Kim, S, Zhang, Z, Upchurch, S, Isern, N, Chen, Y. | | Deposit date: | 2003-12-22 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and DNA-binding sites of the SWI1 AT-rich interaction domain (ARID) suggest determinants for sequence-specific DNA recognition.
J.Biol.Chem., 279, 2004
|
|
6KI9
 
 | | Apo structure of FabMG, novel types of Enoyl-acyl carrier protein reductase | | Descriptor: | 1,2-ETHANEDIOL, FabMG, novel types of Enoyl-acyl carrier protein reductase, ... | | Authors: | Kim, S, Rhee, S. | | Deposit date: | 2019-07-17 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A triclosan-resistance protein from the soil metagenome is a novel enoyl-acyl carrier protein reductase: Structure-guided functional analysis.
Febs J., 287, 2020
|
|
3W9B
 
 | | Crystal structure of Candida antarctica lipase B with anion-tag | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Lipase B | | Authors: | Kim, S.K, Lee, H.H, Park, Y.C, Jeon, S.T, Son, S.H, Min, W.K, Seo, J.H. | | Deposit date: | 2013-04-02 | | Release date: | 2014-04-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Anion tags improve extracellular production of Candida antarctica lipase B in Escherichia coli
To be Published
|
|
1JE4
 
 | |
1L8R
 
 | | Structure of the Retinal Determination Protein Dachshund Reveals a DNA-Binding Motif | | Descriptor: | Dachshund | | Authors: | Kim, S.S, Zhang, R, Braunstein, S.E, Joachimiak, A, Cvekl, A, Hegde, R.S. | | Deposit date: | 2002-03-21 | | Release date: | 2002-06-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the retinal determination protein Dachshund reveals a DNA binding motif.
Structure, 10, 2002
|
|
