7D5V
 
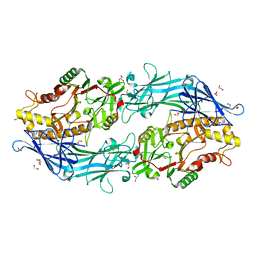 | | Structure of the C646A mutant of peptidylarginine deiminase type III (PAD3) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein-arginine deiminase type-3 | | Authors: | Akimoto, M, Mashimo, R, Unno, M. | | Deposit date: | 2020-09-28 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structures of human peptidylarginine deiminase type III provide insights into substrate recognition and inhibitor design.
Arch.Biochem.Biophys., 708, 2021
|
|
3SSS
 
 | | CcmK1 with residues 103-113 deleted | | Descriptor: | CHLORIDE ION, Carbon dioxide concentrating mechanism protein | | Authors: | Kimber, M.S, Samborska, B. | | Deposit date: | 2011-07-08 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A CcmK2 double layer is the dominant architectural feature of the beta-carboxysomal shell facet
Structure, 2012
|
|
3SSR
 
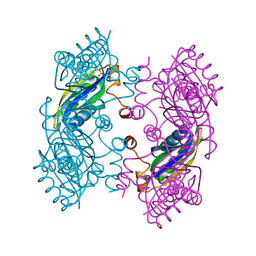 | | CcmK2 dodecamer - form 2 | | Descriptor: | Carbon dioxide concentrating mechanism protein, SULFATE ION | | Authors: | Kimber, M.S, Samborska, B. | | Deposit date: | 2011-07-08 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A CcmK2 double layer is the dominant architectural feature of the beta-carboxysomal shell facet
Structure, 2012
|
|
3EN0
 
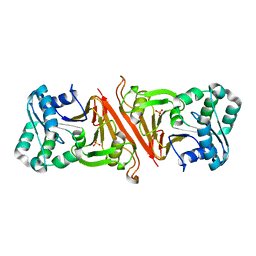 | | The Structure of Cyanophycinase | | Descriptor: | Cyanophycinase, SULFATE ION | | Authors: | Kimber, M.S, Law, A.M, Lai, S.W.S, Tavares, J. | | Deposit date: | 2008-09-25 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structural basis of beta-peptide-specific cleavage by the serine protease cyanophycinase.
J.Mol.Biol., 392, 2009
|
|
2MNG
 
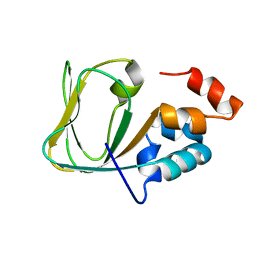 | | Apo Structure of human HCN4 CNBD solved by NMR | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Akimoto, M, Zhang, Z, Boulton, S, Selvaratnam, R, VanSchouwen, B, Gloyd, M, Accili, E.A, Lange, O.F, Melacini, G. | | Deposit date: | 2014-04-03 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A mechanism for the auto-inhibition of hyperpolarization-activated cyclic nucleotide-gated (HCN) channel opening and its relief by cAMP.
J.Biol.Chem., 289, 2014
|
|
3QZY
 
 | | Structure of Baculovirus Sulfhydryl Oxidase Ac92 | | Descriptor: | Baculovirus sulfhydryl oxidase Ac92, FLAVIN-ADENINE DINUCLEOTIDE, IMIDAZOLE, ... | | Authors: | Hakim, M, Fass, D. | | Deposit date: | 2011-03-07 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure of a baculovirus sulfhydryl oxidase, a highly divergent member of the erv flavoenzyme family.
J.Virol., 85, 2011
|
|
3P0K
 
 | | Structure of Baculovirus Sulfhydryl Oxidase Ac92 | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, IMIDAZOLE, ... | | Authors: | Hakim, M, Fass, D. | | Deposit date: | 2010-09-29 | | Release date: | 2011-12-07 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure of a baculovirus sulfhydryl oxidase, a highly divergent member of the erv flavoenzyme family.
J.Virol., 85, 2011
|
|
3GWL
 
 | |
3GWN
 
 | |
3TD7
 
 | | Crysal structure of the mimivirus sulfhydryl oxidase R596 | | Descriptor: | FAD-linked sulfhydryl oxidase R596, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hakim, M, Fass, D. | | Deposit date: | 2011-08-10 | | Release date: | 2012-09-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Exploring ORFan domains in giant viruses: structure of mimivirus sulfhydryl oxidase R596.
Plos One, 7, 2012
|
|
9IKC
 
 | | Orf virus scaffolding protein Orfv075 | | Descriptor: | 62 kDa protein | | Authors: | Hyun, J, Kim, S, Ko, S, Kim, M, Jang, Y. | | Deposit date: | 2024-06-27 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Cryo-EM structure of orf virus scaffolding protein orfv075.
Biochem.Biophys.Res.Commun., 728, 2024
|
|
7XLJ
 
 | | The crystal structure of ORE1(ANAC092) NAC domain | | Descriptor: | NAC domain-containing protein 92 | | Authors: | Chun, I.S, Kim, M.S. | | Deposit date: | 2022-04-21 | | Release date: | 2023-03-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis of DNA binding by the NAC transcription factor ORE1, a master regulator of plant senescence.
Plant Commun., 4, 2023
|
|
8YA8
 
 | |
2PV6
 
 | | HIV-1 gp41 Membrane Proximal Ectodomain Region peptide in DPC micelle | | Descriptor: | Envelope glycoprotein | | Authors: | Sun, Z.-Y.J, Oh, K.J, Kim, M, Reinherz, E.L, Wagner, G. | | Deposit date: | 2007-05-09 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane
Immunity, 28, 2008
|
|
1SIX
 
 | | Mycobacterium tuberculosis dUTPase complexed with magnesium and alpha,beta-imido-dUTP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Sawaya, M.R, Chan, S, Segelke, B, Lekin, T, Krupka, H, Cho, U.-S, Kim, M, So, M, Kim, C.-Y, Naranjo, C.M, Rogers, Y.C, Park, M.S, Waldo, G.S, Pashkov, I, Cascio, D, Yeates, T.O, Perry, J.L, Terwilliger, T.C, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
6CIL
 
 | | PRE-REACTION COMPLEX, RAG1(E962Q)/2-INTACT/INTACT 12/23RSS COMPLEX IN MN2+ | | Descriptor: | High mobility group protein B1, Intact 12RSS substrate forward strand, Intact 12RSS substrate reverse strand, ... | | Authors: | Chuenchor, W, Chen, X, Kim, M.S, Gellert, M, Yang, W. | | Deposit date: | 2018-02-24 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
6CIK
 
 | | Pre-Reaction Complex, RAG1(E962Q)/2-intact/nicked 12/23RSS complex in Mn2+ | | Descriptor: | DNA (5'-D(*AP*TP*CP*TP*GP*GP*CP*CP*TP*GP*TP*CP*TP*TP*A)-3'), High mobility group protein B1, Intact 12RSS substrate forward strand, ... | | Authors: | Chuenchor, W, Chen, X, Kim, M.S, Gellert, M, Yang, W. | | Deposit date: | 2018-02-24 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
6CIM
 
 | | Pre-Reaction Complex, RAG1(E962Q)/2-nicked/intact 12/23RSS complex in Mn2+ | | Descriptor: | DNA (5'-D(*GP*CP*CP*TP*GP*TP*CP*TP*TP*A)-3'), High mobility group protein B1, Intact 23RSS substrate forward strand, ... | | Authors: | Chuenchor, W, Chen, X, Kim, M.S, Gellert, M, Yang, W. | | Deposit date: | 2018-02-24 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
7XP3
 
 | | DNA complex form of ORESARA1(ANAC092) NAC Domain | | Descriptor: | DNA (5'-D(*AP*GP*TP*TP*AP*CP*GP*TP*AP*CP*GP*GP*CP*AP*CP*AP*CP*GP*TP*AP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*TP*AP*CP*GP*TP*GP*TP*GP*CP*CP*GP*TP*AP*CP*GP*TP*AP*AP*C)-3'), DNA (5'-D(P*CP*AP*CP*AP*CP*GP*TP*AP*AP*C)-3'), ... | | Authors: | Chun, I.S, Kim, M.S. | | Deposit date: | 2022-05-03 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural basis of DNA binding by the NAC transcription factor ORE1, a master regulator of plant senescence.
Plant Commun., 4, 2023
|
|
8SLU
 
 | | Crystal structure of human STEP (PTPN5) at cryogenic temperature (100 K) and high pressure (205 MPa) | | Descriptor: | GLYCEROL, SULFATE ION, Tyrosine-protein phosphatase non-receptor type 5 | | Authors: | Ebrahim, A, Guerrero, L, Riley, B.T, Kim, M, Huang, Q, Finke, A.D, Keedy, D.A. | | Deposit date: | 2023-04-24 | | Release date: | 2023-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Pushed to extremes: distinct effects of high temperature vs. pressure on the structure of an atypical phosphatase.
Biorxiv, 2023
|
|
8SLS
 
 | | Crystal structure of human STEP (PTPN5) at cryogenic temperature (100 K) and ambient pressure (0.1 MPa) | | Descriptor: | GLYCEROL, SULFATE ION, Tyrosine-protein phosphatase non-receptor type 5 | | Authors: | Ebrahim, A, Guerrero, L, Riley, B.T, Kim, M, Huang, Q, Finke, A.D, Keedy, D.A. | | Deposit date: | 2023-04-24 | | Release date: | 2023-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Pushed to extremes: distinct effects of high temperature vs. pressure on the structure of an atypical phosphatase.
Biorxiv, 2023
|
|
8SLT
 
 | | Crystal structure of human STEP (PTPN5) at physiological temperature (310 K) and ambient pressure (0.1 MPa) | | Descriptor: | SULFATE ION, Tyrosine-protein phosphatase non-receptor type 5 | | Authors: | Ebrahim, A, Guerrero, L, Riley, B.T, Kim, M, Huang, Q, Finke, A.D, Keedy, D.A. | | Deposit date: | 2023-04-24 | | Release date: | 2023-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Pushed to extremes: distinct effects of high temperature vs. pressure on the structure of an atypical phosphatase.
Biorxiv, 2023
|
|
5JXD
 
 | | Crystal structure of murine Tnfaip8 C165S mutant | | Descriptor: | Tumor necrosis factor alpha-induced protein 8, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Park, J, Kim, M.S, Lee, D, Shin, D.H. | | Deposit date: | 2016-05-13 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.029 Å) | | Cite: | The Tnfaip8-PE complex is a novel upstream effector in the anti-autophagic action of insulin
Sci Rep, 7, 2017
|
|
8W7X
 
 | | SPS_Carbonic Anhydrases | | Descriptor: | 1,2-ETHANEDIOL, BICARBONATE ION, SPS_Carbon Anhydrase, ... | | Authors: | Chun, I.S, Kim, M.S. | | Deposit date: | 2023-08-31 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | structure of SPS_Carbon Anhydrase
To Be Published
|
|
6KYV
 
 | | Crystal Structure of RIG-I and hairpin RNA with G-U wobble base pairs | | Descriptor: | Probable ATP-dependent RNA helicase DDX58, RNA (5'-R(*GP*GP*UP*AP*GP*AP*CP*GP*CP*UP*UP*CP*GP*GP*CP*GP*UP*UP*UP*GP*CP*C)-3'), ZINC ION | | Authors: | Kim, K.-H, Hwang, J, Kim, J.H, Son, K.-P, Jang, Y, Kim, M, Kang, S.-J, Lee, J.-O, Choi, B.-S. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biophysical properties of RIG-I bound to dsRNA with G-U wobble base pairs.
Rna Biol., 17, 2020
|
|
