8J1O
 
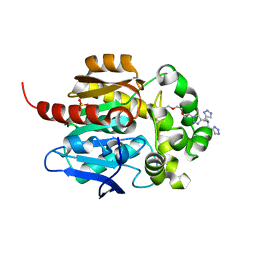 | | Crystal structure of HaloTag complexed with BTTA | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase, N-[[1-[2-[2-(2-hexoxyethoxy)ethoxy]ethyl]-1,2,3-triazol-4-yl]methyl]-1-(1H-1,2,3-triazol-4-yl)-N-(2H-1,2,3-triazol-4-ylmethyl)methanamine, ... | | Authors: | Kim, H, Rhee, H, Lee, C. | | Deposit date: | 2023-04-13 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | Crystal structure of HaloTag complexed with BTTA
To Be Published
|
|
6NBA
 
 | | Crystal structure of Human Cystathionine gamma lyase with S-3-Carboxpropyl-L-Cysteine | | Descriptor: | 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, Cystathionine gamma-lyase | | Authors: | Kim, H, Yadav, P.K, Banerjee, R, Cho, U.-S. | | Deposit date: | 2018-12-06 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | S-3-Carboxypropyl-l-cysteine specifically inhibits cystathionine gamma-lyase-dependent hydrogen sulfide synthesis.
J.Biol.Chem., 294, 2019
|
|
1ZDR
 
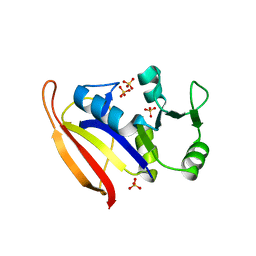 | | DHFR from Bacillus Stearothermophilus | | Descriptor: | GLYCEROL, SULFATE ION, dihydrofolate reductase | | Authors: | Kim, H.S, Damo, S.M, Lee, S.Y, Wemmer, D, Klinman, J.P. | | Deposit date: | 2005-04-14 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and hydride transfer mechanism of a moderate thermophilic dihydrofolate reductase from Bacillus stearothermophilus and comparison to its mesophilic and hyperthermophilic homologues.
Biochemistry, 44, 2005
|
|
2MSY
 
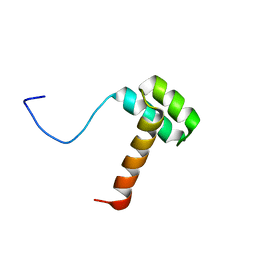 | | Solution structure of Hox homeodomain | | Descriptor: | Homeobox protein Hox-C9 | | Authors: | Kim, H, Park, S, Han, J, Lee, B. | | Deposit date: | 2014-08-11 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insight into the interaction between the Hox and HMGB1 and understanding of the HMGB1-enhancing effect of Hox-DNA binding.
Biochim.Biophys.Acta, 1854, 2015
|
|
1J32
 
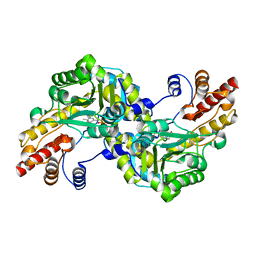 | |
4Q2Y
 
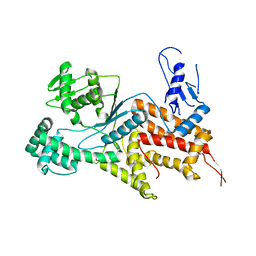 | | Crystal structure of Arginyl-tRNA synthetase | | Descriptor: | Arginine--tRNA ligase, cytoplasmic | | Authors: | Kim, H.S, Jo, C.H, Cha, S.Y, Han, A.R, Hwang, K.Y. | | Deposit date: | 2014-04-10 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | The crystal structure of arginyl-tRNA synthetase from Homo sapiens
Febs Lett., 588, 2014
|
|
4Q2T
 
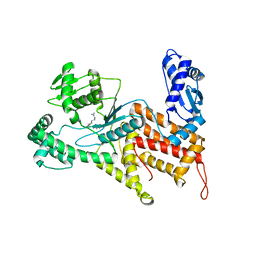 | | Crystal structure of Arginyl-tRNA synthetase complexed with L-arginine | | Descriptor: | ARGININE, Arginine--tRNA ligase, cytoplasmic, ... | | Authors: | Kim, H.S, Jo, C.H, Cha, S.Y, Han, A.R, Hwang, K.Y. | | Deposit date: | 2014-04-09 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of arginyl-tRNA synthetase from Homo sapiens
Febs Lett., 588, 2014
|
|
1GYP
 
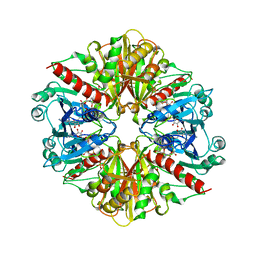 | | CRYSTAL STRUCTURE OF GLYCOSOMAL GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE FROM LEISHMANIA MEXICANA: IMPLICATIONS FOR STRUCTURE-BASED DRUG DESIGN AND A NEW POSITION FOR THE INORGANIC PHOSPHATE BINDING SITE | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Kim, H, Feil, I.K, Verlinde, C.L.M.J, Petra, P.H, Hol, W.G.J. | | Deposit date: | 1995-08-01 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of glycosomal glyceraldehyde-3-phosphate dehydrogenase from Leishmania mexicana: implications for structure-based drug design and a new position for the inorganic phosphate binding site.
Biochemistry, 34, 1995
|
|
4Q2X
 
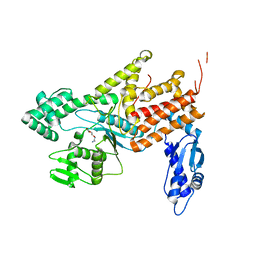 | | Crystal structure of Arginyl-tRNA synthetase complexed with L-canavanine | | Descriptor: | Arginine--tRNA ligase, cytoplasmic, L-CANAVANINE | | Authors: | Kim, H.S, Jo, C.H, Cha, S.Y, Han, A.R, Hwang, K.Y. | | Deposit date: | 2014-04-10 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | The crystal structure of arginyl-tRNA synthetase from Homo sapiens
Febs Lett., 588, 2014
|
|
5X90
 
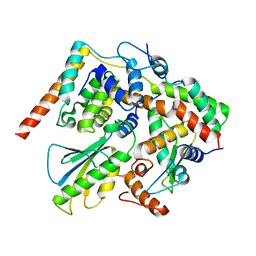 | | Structure of DotL(656-783)-IcmS-IcmW-LvgA derived from Legionella pneumophila | | Descriptor: | Hypothetical virulence protein, IcmO (DotL), IcmS, ... | | Authors: | Kim, H, Kwak, M.J, Kim, J.D, Kim, Y.G, Oh, B.H. | | Deposit date: | 2017-03-04 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Architecture of the type IV coupling protein complex of Legionella pneumophila
Nat Microbiol, 2, 2017
|
|
7VWX
 
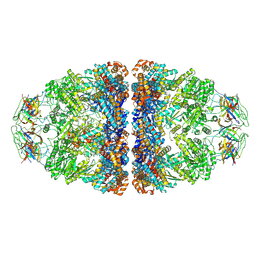 | | CryoEM structure of football-shaped GroEL:ES2 with RuBisCO | | Descriptor: | Chaperonin GroEL, Co-chaperonin GroES, Ribulose bisphosphate carboxylase | | Authors: | Kim, H, Roh, S.H. | | Deposit date: | 2021-11-12 | | Release date: | 2022-01-12 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Cryo-EM structures of GroEL:ES 2 with RuBisCO visualize molecular contacts of encapsulated substrates in a double-cage chaperonin.
Iscience, 25, 2022
|
|
7X15
 
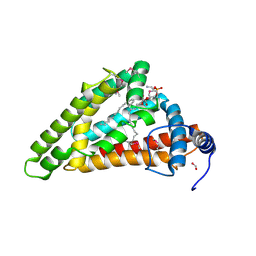 | | Crystal structure of MIGA2 LD targeting domain | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, FORMIC ACID, Mitoguardin 2 | | Authors: | Kim, H, Lee, C. | | Deposit date: | 2022-02-23 | | Release date: | 2022-09-14 | | Method: | X-RAY DIFFRACTION (2.852 Å) | | Cite: | Structural basis for mitoguardin-2 mediated lipid transport at ER-mitochondrial membrane contact sites.
Nat Commun, 13, 2022
|
|
7X14
 
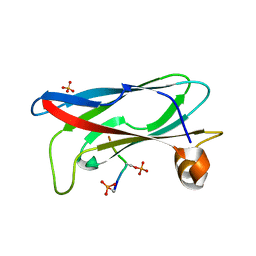 | | Crystal structure of phospho-FFAT motif of MIGA2 bound to VAPB | | Descriptor: | MIGA2 phospho FFAT motif, SULFATE ION, Vesicle-associated membrane protein-associated protein B | | Authors: | Kim, H, Lee, C. | | Deposit date: | 2022-02-23 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.675 Å) | | Cite: | Structural basis for mitoguardin-2 mediated lipid transport at ER-mitochondrial membrane contact sites.
Nat Commun, 13, 2022
|
|
3OUH
 
 | | PHD2-R127 with JNJ41536014 | | Descriptor: | 1-(5-chloro-6-fluoro-1H-benzimidazol-2-yl)-1H-pyrazole-4-carboxylic acid, Egl nine homolog 1, FE (II) ION, ... | | Authors: | Kim, H, Clark, R. | | Deposit date: | 2010-09-14 | | Release date: | 2010-12-01 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Benzimidazole-2-pyrazole HIF Prolyl 4-Hydroxylase Inhibitors as Oral Erythropoietin Secretagogues.
ACS Med Chem Lett, 1, 2010
|
|
8HEJ
 
 | | Crystal structure of Transthyretin in complex with a covalent inhibitor trans-styrylpyrazole | | Descriptor: | 2,4,6-trifluorobenzaldehyde, 2,6-dibromo-4-[(E)-2-(3,5-dimethyl-1H-pyrazol-4-yl)ethenyl]phenol, Transthyretin | | Authors: | Kim, H, Choi, S, Lee, C. | | Deposit date: | 2022-11-08 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of Transthyretin in complex with a covalent inhibitor trans-styrylpyrazole
To Be Published
|
|
4O6G
 
 | | Rv3902c from M. tuberculosis | | Descriptor: | Uncharacterized protein | | Authors: | Reddy, B.G, Moates, D.B, Kim, H, Green, T.J, Kim, C, Terwilliger, T.J, Delucas, L.J, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2013-12-20 | | Release date: | 2014-03-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55 angstrom resolution X-ray crystal structure of Rv3902c from Mycobacterium tuberculosis.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4UIP
 
 | | The complex structure of extracellular domain of EGFR with Repebody (rAC1). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, EPIDERMAL GROWTH FACTOR RECEPTOR, ... | | Authors: | Kang, Y.J, Cha, Y.J, Cho, H.S, Lee, J.J, Kim, H.S. | | Deposit date: | 2015-03-31 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Enzymatic Prenylation and Oxime Ligation for the Synthesis of Stable and Homogeneous Protein-Drug Conjugates for Targeted Therapy.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7CTP
 
 | |
4ZG0
 
 | | Crystal structure of Mouse Syndesmos protein | | Descriptor: | Protein syndesmos | | Authors: | Lee, I, Kim, H, Yoo, J, Cho, H, Lee, W. | | Deposit date: | 2015-04-22 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Crystal structure of syndesmos and its interaction with Syndecan-4 proteoglycan
Biochem.Biophys.Res.Commun., 463, 2015
|
|
8GTG
 
 | | Corticotropin-releasing hormone receptor 1(CRF1R) bound with BMK-I-152 by XFEL | | Descriptor: | 8-(4-bromanyl-2,6-dimethoxy-phenyl)-~{N},~{N}-bis(2-methoxyethyl)-2,7-dimethyl-pyrazolo[1,5-a][1,3,5]triazin-4-amine, Endolysin, Isoform CRF-R2 of Corticotropin-releasing factor receptor 1 | | Authors: | Cho, H.S, Kim, H. | | Deposit date: | 2022-09-08 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-based drug discovery of a corticotropin-releasing hormone receptor 1 antagonist using an X-ray free-electron laser.
Exp.Mol.Med., 55, 2023
|
|
8GTI
 
 | | Corticotropin-releasing hormone receptor 1(CRF1R) bound with BMK-C205 by XFEL | | Descriptor: | 8-(4-bromanyl-2,6-dimethoxy-phenyl)-~{N}-butyl-~{N}-(cyclopropylmethyl)-2,7-dimethyl-pyrazolo[1,5-a][1,3,5]triazin-4-amine, Endolysin, Isoform CRF-R2 of Corticotropin-releasing factor receptor 1, ... | | Authors: | Cho, H.S, Kim, H. | | Deposit date: | 2022-09-08 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based drug discovery of a corticotropin-releasing hormone receptor 1 antagonist using an X-ray free-electron laser.
Exp.Mol.Med., 55, 2023
|
|
8GTM
 
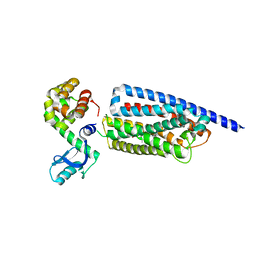 | | Corticotropin-releasing hormone receptor 1(CRF1R) bound with BMK-C203 by XFEL | | Descriptor: | 7-(4-bromanyl-2,6-dimethoxy-phenyl)-4,8-dimethyl-~{N},~{N}-bis[4,4,4-tris(fluoranyl)butyl]-1$l^{4},3,5,9-tetrazabicyclo[4.3.0]nona-1(6),2,4,8-tetraen-2-amine, Endolysin, Isoform CRF-R2 of Corticotropin-releasing factor receptor 1 | | Authors: | Cho, H.S, Kim, H. | | Deposit date: | 2022-09-08 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based drug discovery of a corticotropin-releasing hormone receptor 1 antagonist using an X-ray free-electron laser.
Exp.Mol.Med., 55, 2023
|
|
5GLB
 
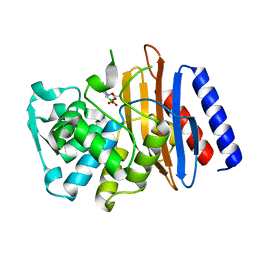 | | Crystal structure of the class A beta-lactamase PenL-tTR10 in complex with CBA | | Descriptor: | Beta-lactamase, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE | | Authors: | Choi, J.M, Yi, H, Kim, H.S, Lee, S.H. | | Deposit date: | 2016-07-10 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High adaptability of the omega loop underlies the substrate-spectrum-extension evolution of a class A beta-lactamase, PenL
Sci Rep, 6, 2016
|
|
4FYB
 
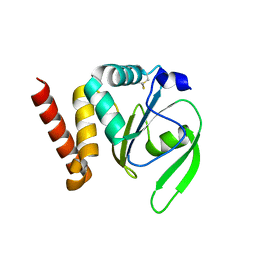 | | Structural and functional characterizations of a thioredoxin-fold protein from Helicobacter pylori | | Descriptor: | GLYCEROL, Thiol:disulfide interchange protein (DsbC) | | Authors: | Yoon, J.Y, Kim, J, Lee, S.J, Im, H.N, Kim, H.S, Yoon, H, An, D.R, Kim, J.Y, Kim, S, Han, B.W, Suh, S.W. | | Deposit date: | 2012-07-04 | | Release date: | 2013-05-08 | | Last modified: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional characterization of HP0377, a thioredoxin-fold protein from Helicobacter pylori
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4FYC
 
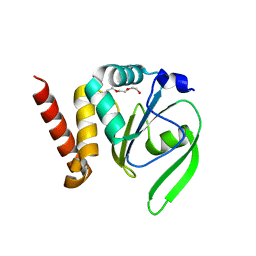 | | Structural and functional characterizations of a thioredoxin-fold protein from Helicobacter pylori | | Descriptor: | TETRAETHYLENE GLYCOL, Thiol:disulfide interchange protein (DsbC) | | Authors: | Yoon, J.Y, Kim, J, Lee, S.J, Im, H.N, Kim, H.S, Yoon, H, An, D.R, Kim, J.Y, Kim, S, Han, B.W, Suh, S.W. | | Deposit date: | 2012-07-04 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural and functional characterization of HP0377, a thioredoxin-fold protein from Helicobacter pylori
Acta Crystallogr.,Sect.D, 69, 2013
|
|
